 Reporter Cell Lines
Reporter Cell Lines
Reporter cell lines are functional cell models used to reflect intracellular signaling changes and gene transcriptional activity in real-time and quantitatively by introducing specific reporter genes. These cell lines are widely used in fields such as gene expression regulation research, signaling pathway analysis, protein localization tracking, receptor-ligand interaction analysis, as well as high-throughput drug screening and new drug target validation. They are indispensable tools in modern molecular and cellular biology research.
EDITGENE, relying on its mature gene overexpression experimental systems and gene knock-in technology platforms, has constructed various stable reporter cell lines. These cover multiple classic signaling pathways and popular tumor therapeutic targets, such as NF-κB, NFAT, GPCR, and cAMP pathways, and genes like EGFR, PIK3CA, and KRAS. The systems utilize various reporter genes or tags, including GFP, RFP, Luciferase, and HIBIT, to meet the needs of diverse experimental platforms and research directions.
In-Stock stable cell lines
Service Details
| Cell Types | Various types including tumor cell lines, normal somatic cell lines, stem cells, and primary cells. |
|---|---|
| Services | Reporter genes or tags such as GFP, RFP, Luciferase, HIBIT, etc. |
| Delivery Standard | 1 gene overexpression polyclonal cell line (2 tubes, 1×10^6 cells/tube) / 1 gene knock-in monoclonal cell line (2 tubes, 1×10^6 cells/tube). |
| Turnaround / Price |
|
Technical Principle
Overexpression reporter cell lines are created by placing a reporter gene under the control of a specific promoter or signaling pathway response element, which is then stably integrated into the cell genome. These cell lines feature sensitive signal response, a wide detection window, and high construction efficiency. They are ideal for signaling pathway activation/inhibition studies, high-throughput drug screening, and functional primary screening.
Utilizing gene knock-in technology platforms such as HES-KI and Flash-KI, knock-in reporter cell lines precisely insert the reporter gene into endogenous gene loci or their regulatory regions. This allows the reporter signal to be generated within the endogenous expression context, providing a more authentic reflection of gene regulation and signaling changes under physiological conditions. These are suitable for mechanistic research, detailed functional analysis, and result validation experiments.
Case Study
K562-EGFP Reporter Cell Lines:EGFP Gene Knock-in Mediated by AsCas12a Using the HES-KI System
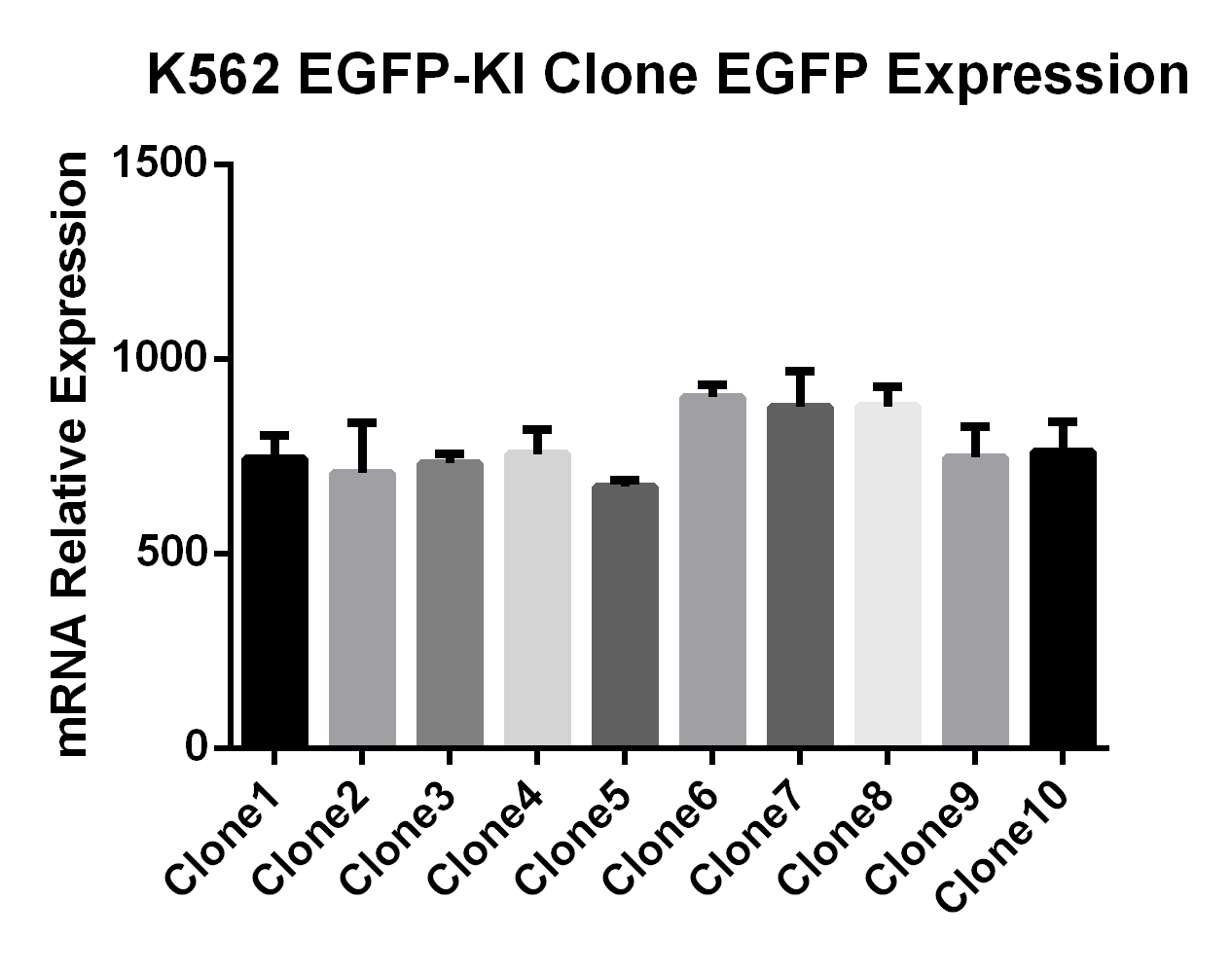
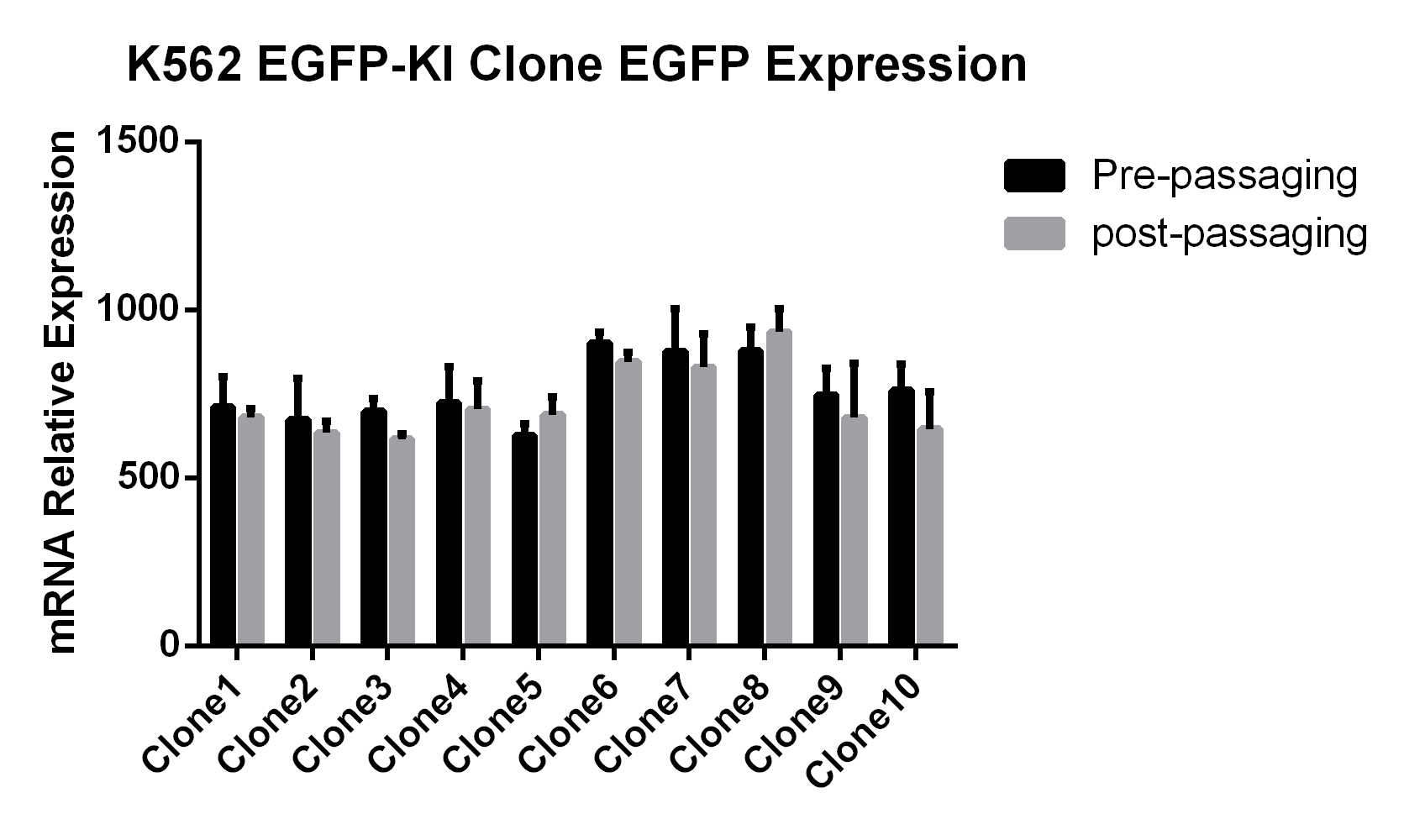
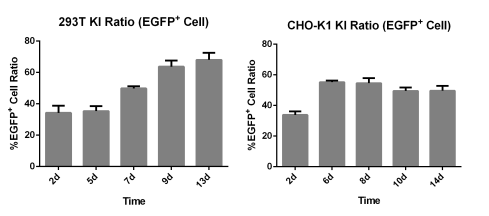
CHO-K1-EGFP Reporter Cell Lines/293T-EGFP Reporter Cell Lines:CRISPR/Cas9-Mediated EGFP Knock-In Based on the HES-KI System
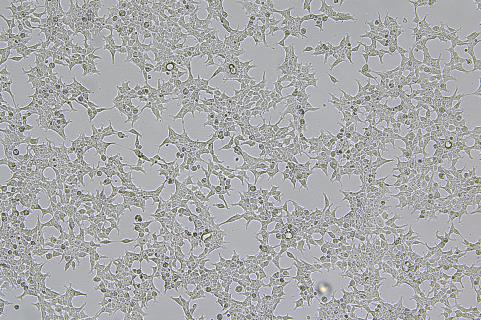
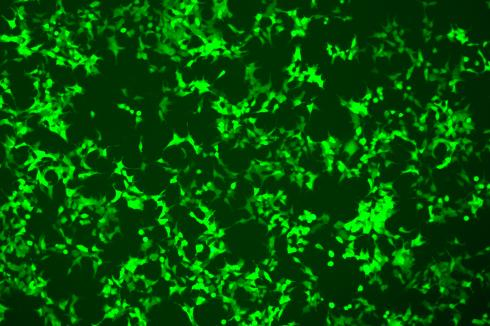
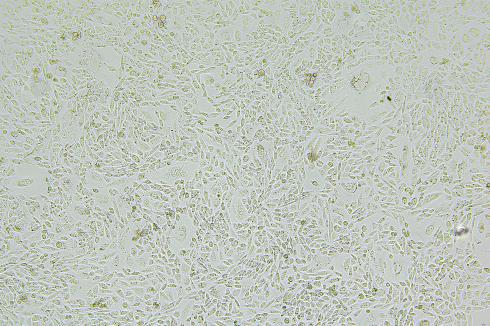
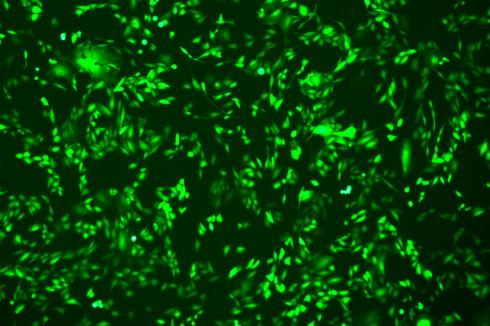
Lentivirus-Mediated Overexpression of copEGFP in A375 Cells
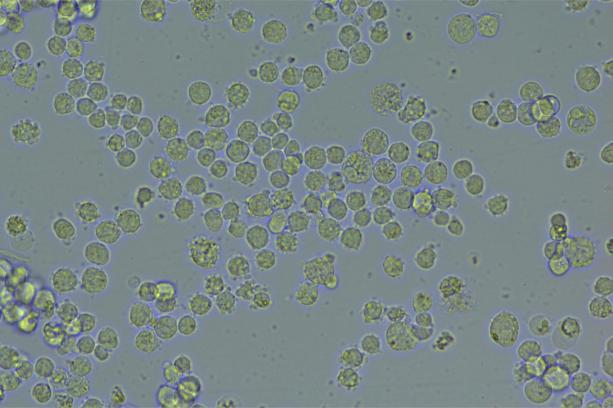
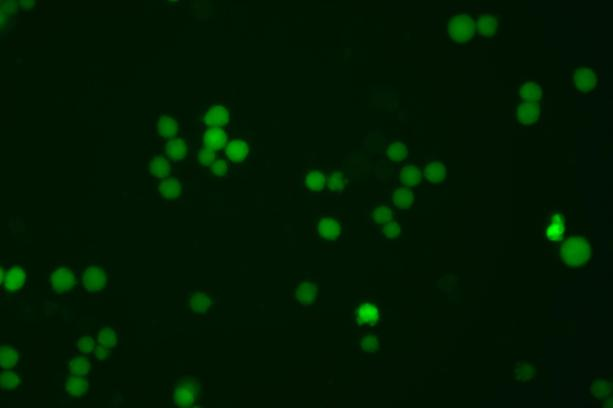
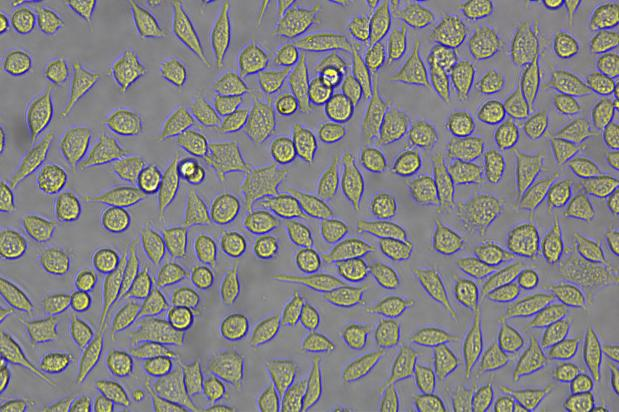
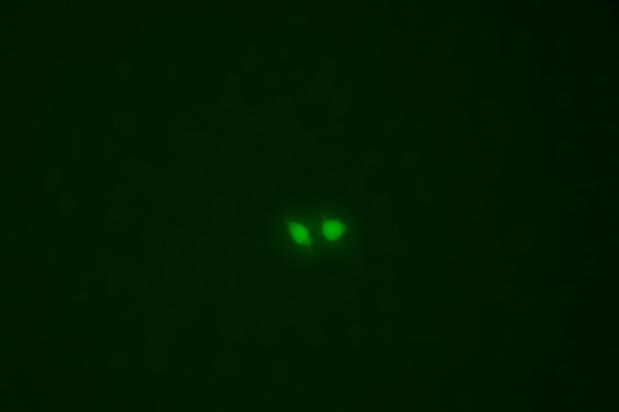
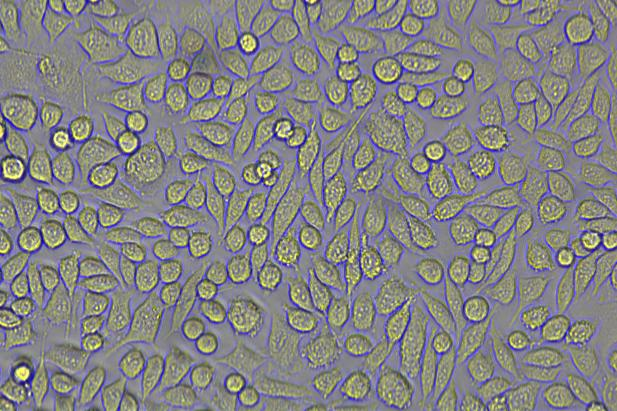
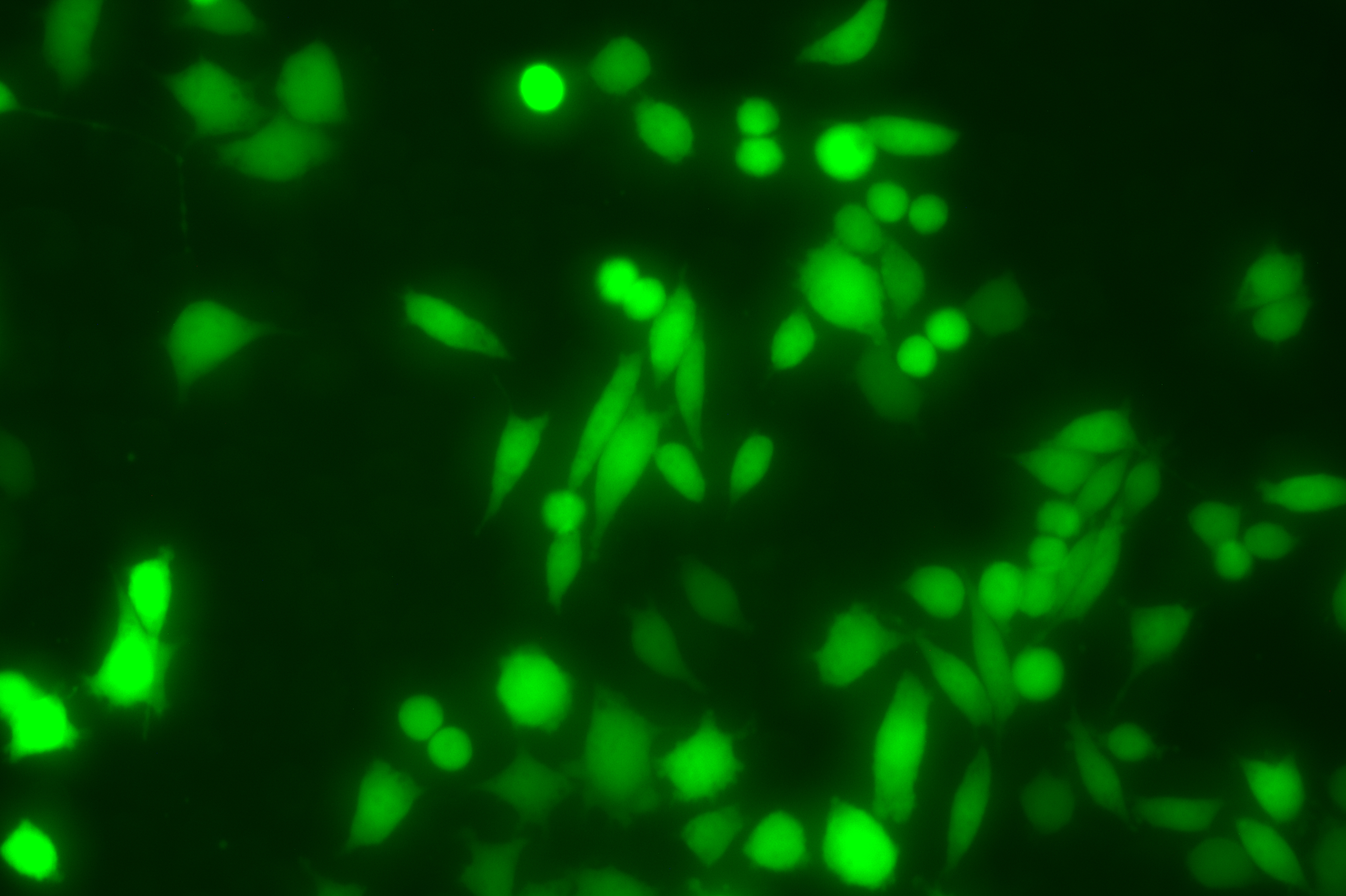
The copEGFP gene sequence was cloned into the lentiviral expression vector pLV3-CAG-MCS-P2A-Puro, followed by packaging and production of high-titer lentiviral particles. These particles were used to infect the human melanoma cell line A375. Expression of the EGFP reporter gene in infected cells was directly observed using fluorescence microscopy to evaluate overexpression efficiency.
Note: TheA375-copEGFP cellsused in this case are ready-to-use stocks provided by EDITGENE (Catalog No.: EDC01053), ensuring rapid experimental setup and reproducibility.copGFP Overexpression Cell Line Stock Library
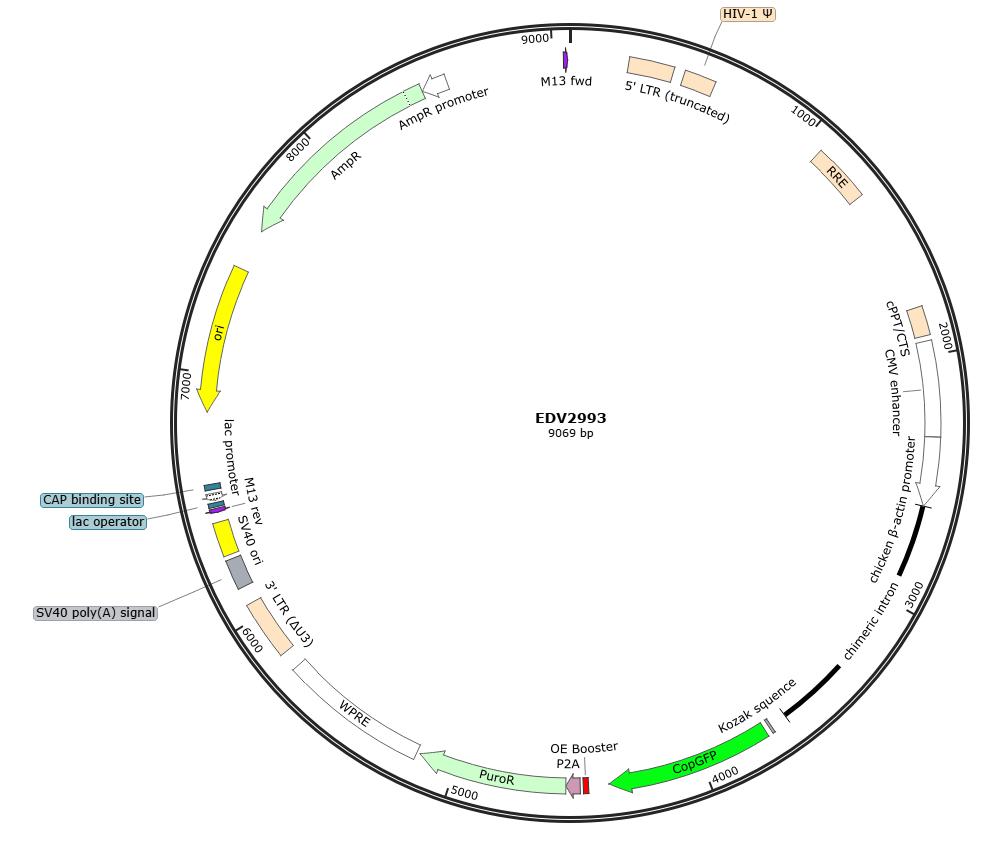
Figure 1: Map of the pLV3-CAG-MCS-P2A-Puro backbone vector
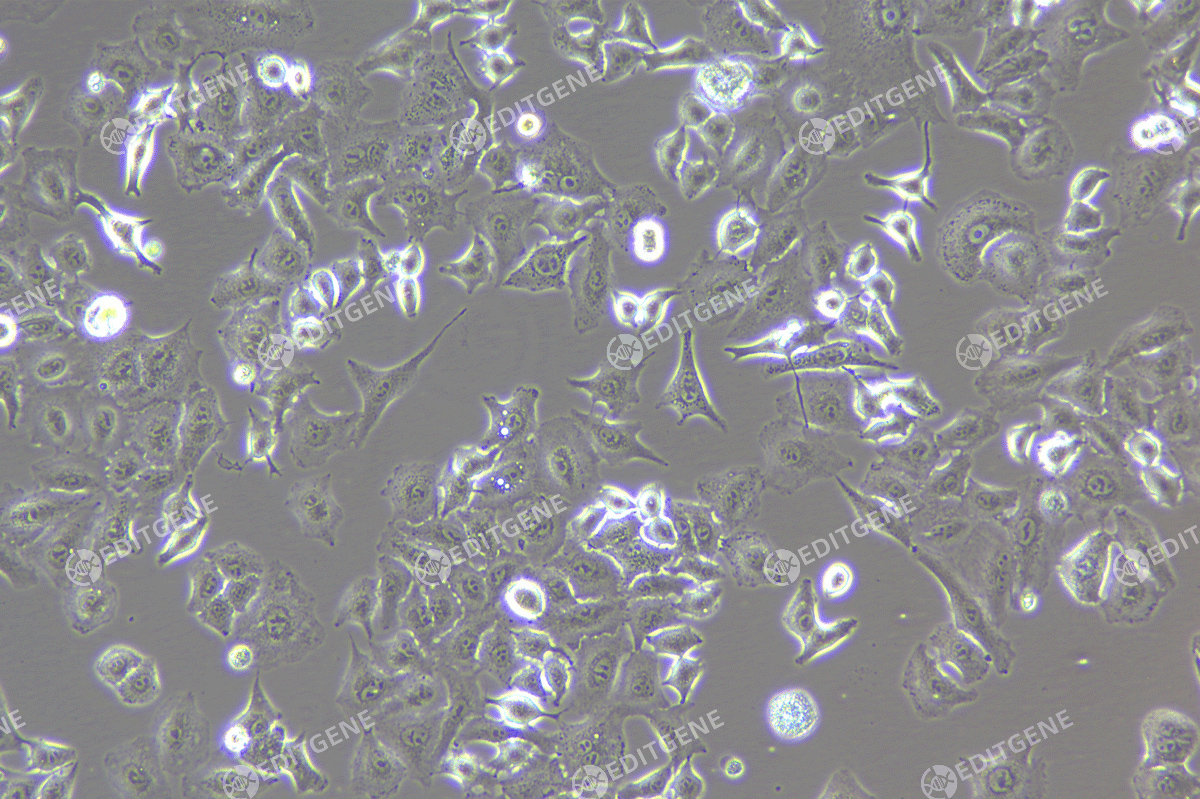
Figure 2: Bright-field image of A375 cells
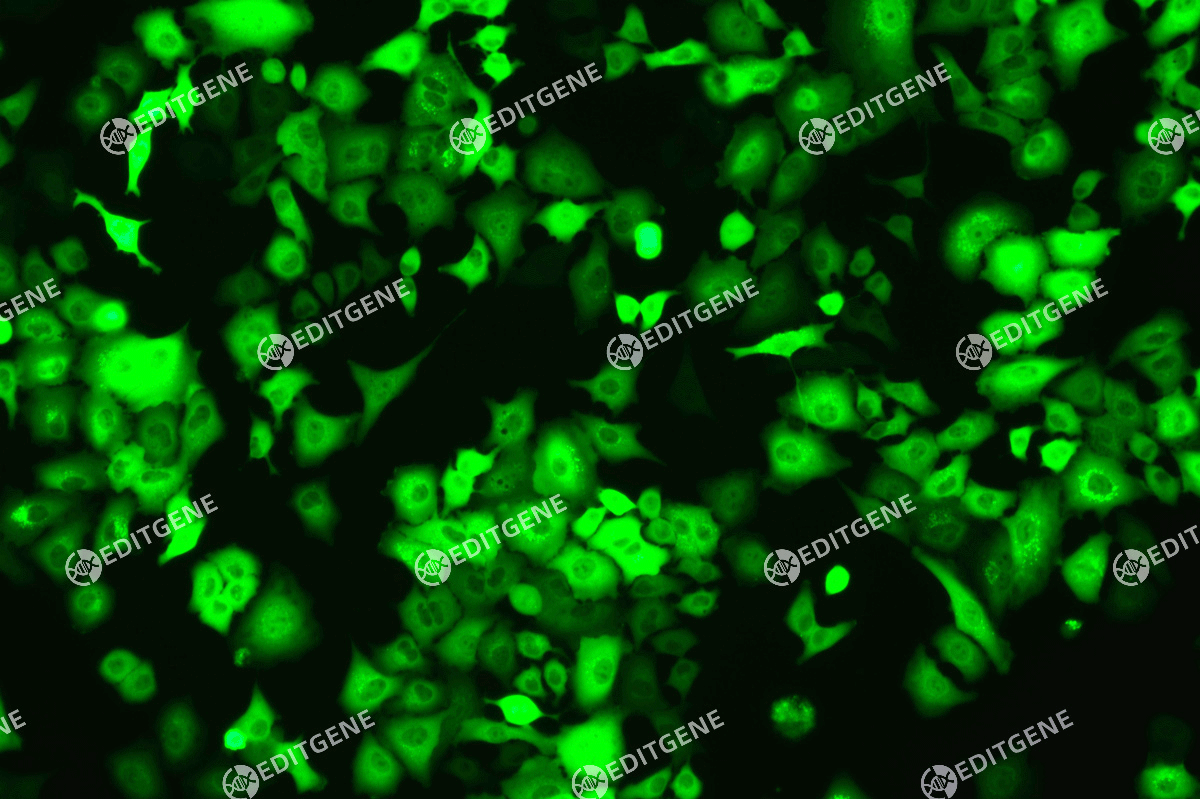
Figure 3: Fluorescence image of A375-copEGFP cells
Precise EGFP Knock-in at Key Gene Loci in HEK293T Cells via Flash-KI System
Using EDITGENE's proprietary Flash-KI platform, the EGFP reporter gene was precisely knocked into the C-terminus of the GAPDH gene in HEK293T cells. Without antibiotic selection, the knock-in efficiency in polyclonal cells reached 88%. Compared to the HES-KI system, the Flash-KI system mediated a 20% increase in knock-in efficiency for critical gene loci.


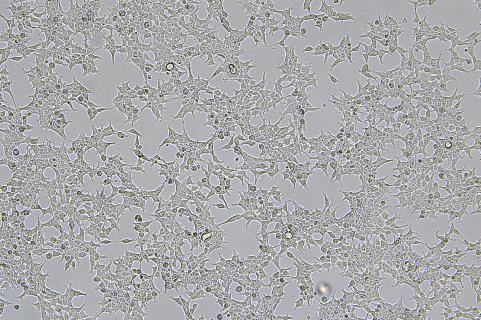
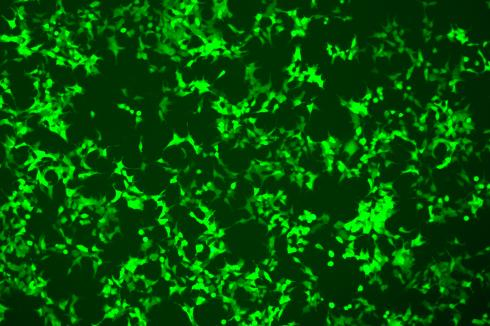
200-300 Fold Efficiency Breakthrough: Precise Knock-in Efficiency in A9 Cells Increased from 0.2% to 46%
In this case, the Flash-KI platform was used to precisely knock the EGFP reporter gene into the C-terminus of the T1R1 gene in A19 cells. Under conditions without antibiotic selection, conventional CRISPR/HDR methods yielded a knock-in efficiency of only 0.2%, consistent with the <1% efficiency typically seen in hard-to-edit cell lines. However, the Flash-KI system increased this efficiency to 46%—a 230-fold improvement.
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Reference Materials
Knock-in strategy based on CLASH technology
The CLASH (Cas9-Linked Adaptor Synthesis for Homology-directed repair) technology enables efficient large-scale gene knock-in for cell engineering. This method combines the Cas9 protein and adaptor synthesis, allowing parallel knock-in across various cell types. By providing specific adaptors during the DNA repair process, it significantly enhances homology-directed repair (HDR) efficiency, thereby increasing knock-in success rates. This technology shows great potential in cell engineering and gene editing, especially for complex bioengineering applications requiring multi-gene modifications.
Enhancing CRISPR-mediated homology-directed repair (HDR) efficiency through cell cycle synchronization
This study explores a method to enhance CRISPR-mediated HDR efficiency by synchronizing the cell cycle. Using small molecules to modulate the cell cycle, researchers achieved a 1.2- to 1.5-fold increase in knock-in efficiency across various cell lines. The study also demonstrated this approach's application in animal embryos, significantly increasing knock-in frequency in pig embryos. This technique improves knock-in success by guiding cells to an HDR-favorable cycle stage, offering a new optimization strategy for CRISPR gene editing.




 Cas9
GFP-LUC Cell Lines
Cas9
GFP-LUC Cell Lines CopGFP Cell Lines
CopGFP Cell Lines
 Luciferase Cell
Lines
Luciferase Cell
Lines