The Key to Enhancing CRISPR Gene Editing Efficiency

CRISPR-Cas9 gene editing technology utilizes custom-designed sgRNA (single-guide RNA) to recognize target genomic sequences and guide the Cas9 nuclease to effectively cleave DNA double strands, creating double-strand breaks. The subsequent repair process results in gene knockouts or knock-ins, ultimately achieving the objective of modifying genomic DNA.
Matthew H. Porteus and colleagues published "Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells" in Nature Biotechnology. This study enhanced the genome editing efficiency in primary human T cells and CD34+ hematopoietic stem and progenitor cells (HSPCs) by incorporating chemical modifications into synthetic sgRNAs. The co-delivery of chemically modified sgRNAs with Cas9 mRNA or protein represents an effective delivery method that avoids the toxicity associated with DNA delivery. This is a simple yet effective approach with the potential to facilitate the development of CRISPR-Cas technology in biotechnology and therapeutic applications.
To test the utility of chemically synthesized sgRNAs for genome editing, the researchers applied three distinct chemical modifications—M, MS, and MSP—to the three nucleotides at both the 5' and 3' termini and evaluated their final performance. The researchers selected three human genes previously targeted by sgRNAs with high gene-editing frequencies in cell lines: IL2RG, HBB, and CCR5. The results indicated that, under normal conditions, the chemically modified sgRNAs maintained high specificity. Differences in the on-target to off-target ratios suggest that chemical alterations to sgRNAs possess the potential to modulate activity.
To further explore chemically modified sgRNAs in human cell lines, the researchers transitioned to an all-RNA delivery platform, co-delivering sgRNA with mRNA encoding Cas9. By utilizing nucleofection for the combined or sequential delivery of Cas9 and various synthetic sgRNAs targeting IL2RG, they investigated whether chemical modifications affect the half-life of sgRNA activity. Subsequently, the researchers compared the activities of unmodified and MS-modified sgRNAs at off-target sites. The study found that when chemically modified sgRNAs were co-delivered with Cas9 mRNA or as RNPs, they demonstrated superior gene editing performance in human cell lines compared to unmodified sgRNAs.
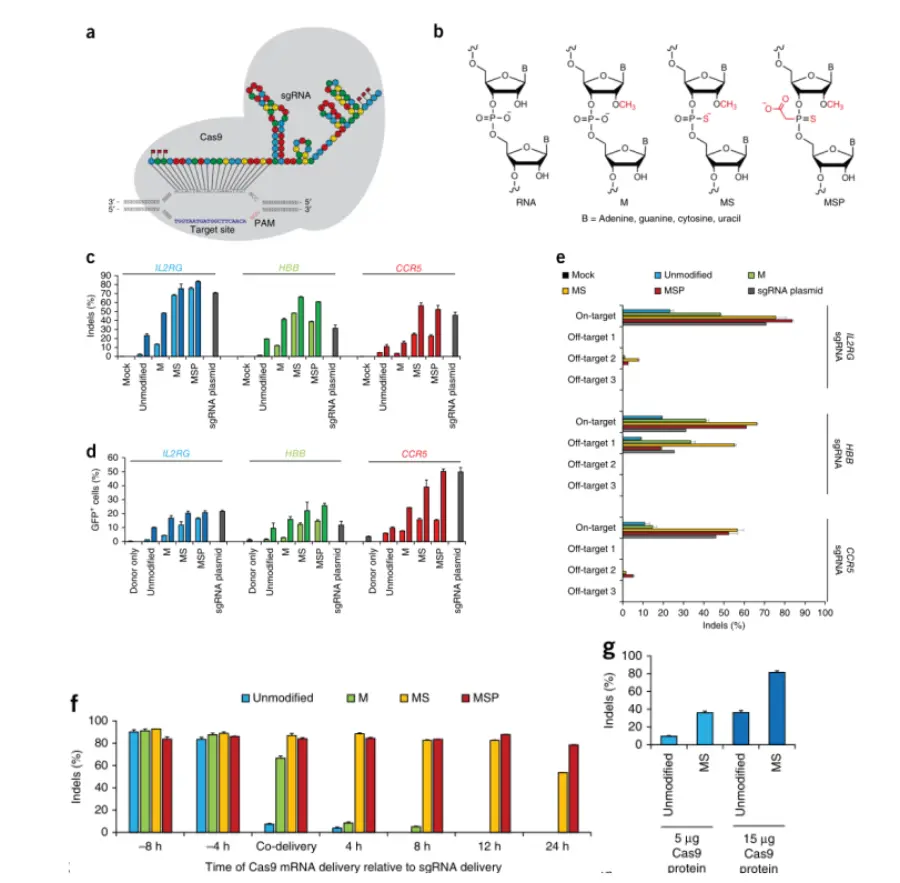
Figure 1. Chemically modified synthetic sgRNAs promote high frequencies of indels and homologous recombination (HR) in the human cell line K562
Next, the researchers tested the chemically modified sgRNAs in primary cells and found that, compared to plasmid nucleofection, the modified sgRNAs had minimal impact on cell survival and proliferation. They also tested MS-modified sgRNAs in unstimulated T cells, which exhibited higher variability between donors compared to stimulated T cells. Furthermore, they discovered that chemically modified sgRNAs targeting IL2RG and HBB were active in CD34+ HSPCs isolated from mobilized peripheral blood.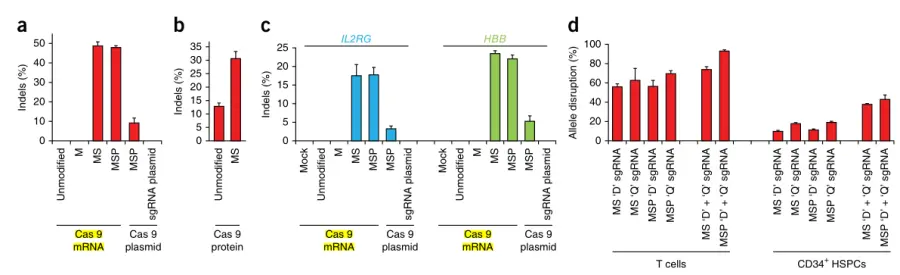
Figure 2. Chemically modified sgRNAs promote high-frequency gene cleavage in human T cells and CD34+ hematopoietic stem and progenitor cells (HSPCs)
In this study, the researchers demonstrated that chemically synthesized sgRNAs can be effectively utilized for targeted genome editing and proved that chemically modified sgRNAs significantly enhance genome editing efficiency in primary human T cells and CD34+ HSPCs. Chemically synthesized and modified sgRNAs offer several advantages over expressed or in vitro transcribed (IVT) sgRNAs, including:
(1)Enhanced therapeutic efficacy.
(2)Stable and sustainable production of high-purity sgRNAs for biotechnological and therapeutic applications.
(3)Greater flexibility in sgRNA design, compared to the constraints imposed on the first transcribed nucleotide by U6 or T7 promoters typically used for plasmid expression or in vitro transcription of sgRNAs.
(4)Lower cytotoxicity in primary cells compared to DNA plasmid-based systems, enabling a high-activity, RNA-only or RNP-based CRISPR platform.
















