How to Design Highly Efficient Cas12a-crRNA Primers

A recent publication by the Marraffini team in Molecular Cell (https://marraffini.rockefeller.edu/) introduced optimized strategies for designing Cas13a-crRNA. For those working on Cas13a detection, this is an invaluable resource for study and application.
Currently, Cas12a is more widely applied in DNA detection. Today, we will focus on learning how to design highly efficient Cas12a-crRNA primers. The following discussion centers on an article published in Nature, specifically addressing the selection of suboptimal PAMs in crRNA primer design. This literature is of great significance for resolving various bottlenecks encountered in traditional "one-tube" CRISPR detection assays.
The Protospacer Adjacent Motif (PAM) is critical for the recognition between the crRNA primer and the target sequence. This is because the Cas12a protein initiates dsDNA target recognition by identifying the PAM sequence, which subsequently facilitates the unwinding of the dsDNA. The canonical PAM sequence is TTTV, located at the 5' end of the crRNA target site.
For newcomers who are just being introduced to CRISPR technology, the role of the PAM might not be fully clear. However, the PAM motif is absolutely vital for CRISPR/Cas-based detection. This article focuses specifically on the study of the PAM motif, where the authors describe a Cas12a-based methodology named sPAMC (suboptimal Protospacer Adjacent Motif for Cas12a-based tests).
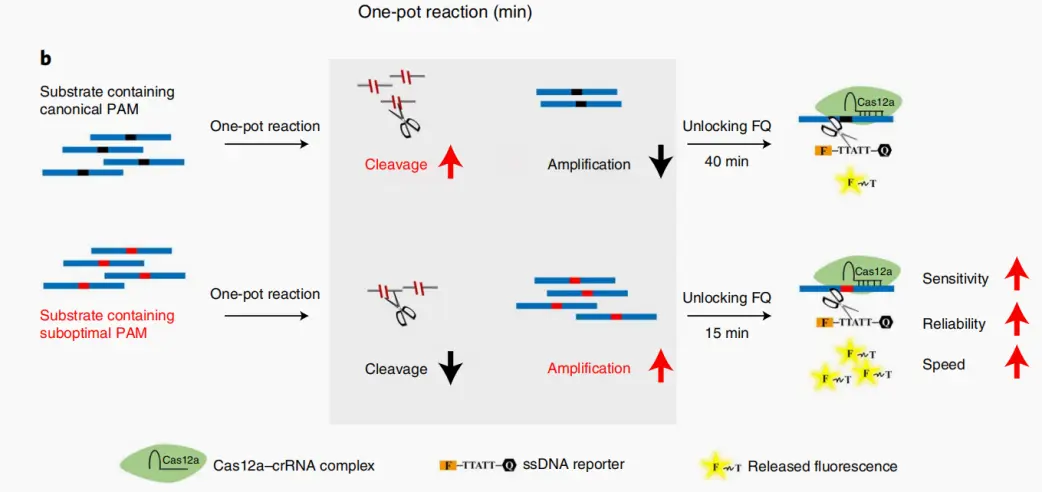
In a "one-tube" CRISPR assay, the isothermal amplification and the Cas12a cleavage reaction occur simultaneously in a single tube. Utilizing CRISPR RNAs (crRNAs) that target suboptimal PAMs instead of canonical PAMs can accelerate the reaction speed by 2 to 3 times. Furthermore, extensive testing demonstrates that sPAMC exhibits significantly higher sensitivity and reliability in one-pot assays compared to those using canonical PAMs.
The term "suboptimal PAM" is used in contrast to the canonical PAM (TTTV). The authors discovered that crRNAs delivering superior performance targeted suboptimal PAMs (such as NTTV and TTNT) rather than the typical TTTV. They further identified that these suboptimal PAMs are the key factor in enhancing Cas12a-mediated detection in one-tube reactions.
Subsequently, the authors explored which specific types of suboptimal PAMs yield the highest sensitivity. By introducing point mutations into the L1 gene of Human Papillomavirus Type 18 (HPV18)—changing TTTV to VTTV, TVTV, or TTVV—they concluded through extensive experimentation that most PAMs involving VTTV, TCTV, and TTVV, as well as some TRTV and TTNT (excluding TTTV), outperform the canonical TTTV motif in one-pot reactions.
Through a series of validation experiments, the researchers confirmed the impact of suboptimal PAMs on detection sensitivity, further solidifying their essential role in optimizing one-tube CRISPR assays.
The underlying principle lies in the competition for the target template within the one-tube CRISPR reaction, where isothermal amplification and Cas enzyme cleavage occur simultaneously. Both processes require the target DNA, creating a competitive relationship.
Using a suboptimal PAM slows down the Cas enzyme's recognition of the target, thereby moderating the rate of the cleavage reaction. This allows the overall reaction kinetics to favor the amplification stage, ensuring that the target can be amplified sufficiently to be detected even at extremely low initial concentrations.
CRISPR Detection Schematic
This article introduces the performance of suboptimal PAMs in CRISPR detection and explores which specific PAM sites yield optimal results through experimental research. While the specific design should be tailored to individual experimental conditions, this method is highly recommended for designing crRNA primers to ensure maximum sensitivity in one-pot reactions. Notably, in two-step (two-tube) reaction formats, suboptimal PAMs still perform less effectively than the canonical PAM structure.
In addition to considering suboptimal PAMs, the design of Cas12a-crRNA primers must also account for multiple factors, including spacer sequence selection and primer secondary structures, to achieve a comprehensive optimization.
















