A Clear Guide to the Principles of CRISPR Detection Technology

CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, which describes a structure where identical short sequences are inserted uniformly across a stretch of DNA, much like organized markers within a sequence of disordered DNA. These identical segments are known as "repeats," while the variable DNA sequences that separate them are called "spacers." Cas stands for CRISPR-associated, and together they are referred to as the CRISPR/Cas system.
The CRISPR/Cas system originates from the bacterial immune system. In prokaryotes, the CRISPR-Cas system functions as an adaptive immune defense, utilizing CRISPR RNAs (crRNAs) as guide molecules to target and recognize invading nucleic acids, thereby defending the cell against viral intrusion.
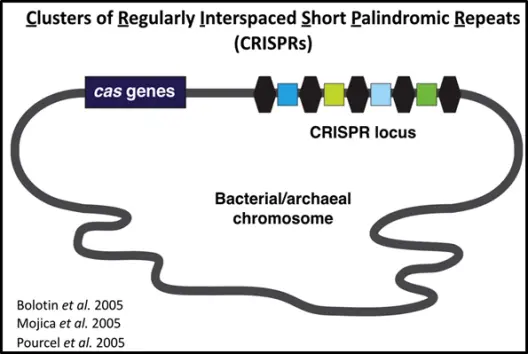
Figure 1. The CRISPR/Cas System
The process primarily consists of the following three stages:
1.Adaptation Stage: DNA fragments from invading viruses or bacteriophages are integrated into the CRISPR array as "spacers."
2.Expression Stage: The CRISPR array is transcribed into a primary transcript (pre-crRNA), which is then processed by Cas proteins to generate mature CRISPR RNA (crRNA).
3.Interference Stage: Mature crRNA complexed with Cas proteins forms a ribonucleoprotein (RNP) complex. Guided by the crRNA, the Cas protein specifically targets and cleaves the nucleic acids of the invader to achieve immune defense.
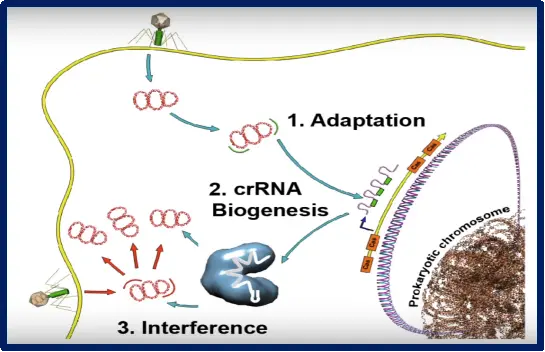
Figure 2. Three distinct mechanistic stages of CRISPR-Cas immunity
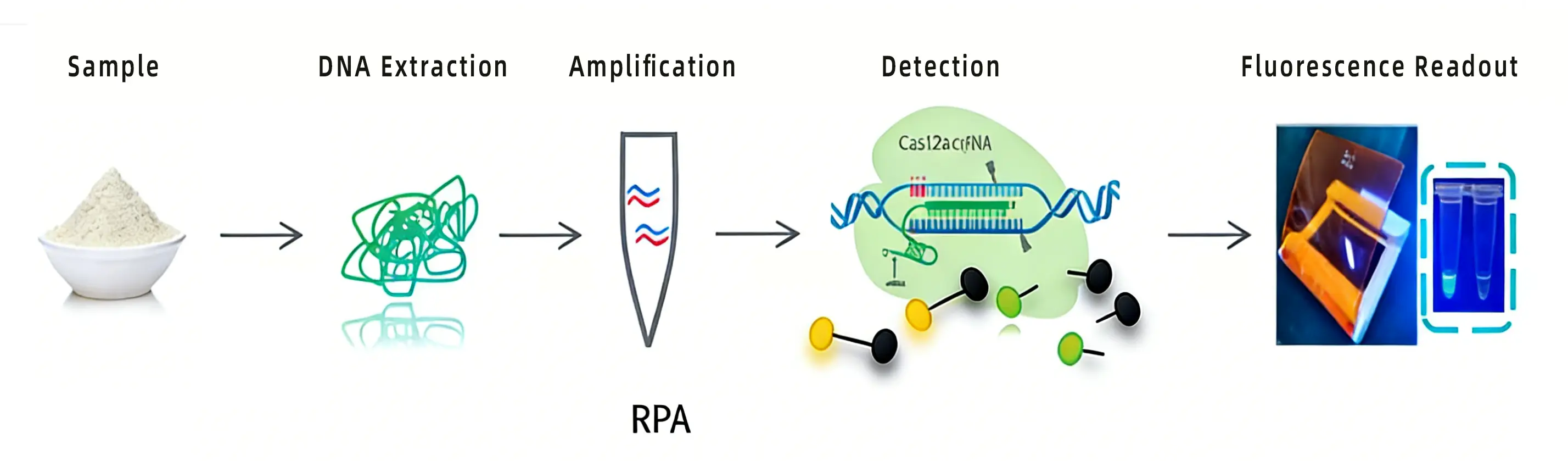
With an understanding of the origins of CRISPR/Cas and its role in bacterial immunity, researchers began applying this system to gene editing. Now, let’s explore how the CRISPR/Cas system is utilized in CRISPR detection.
The primary Cas proteins used in CRISPR detection are Cas12, Cas13, and Cas14. Unlike Cas9, these proteins possess a unique functional capability: in addition to specifically cleaving a target sequence, they exhibit non-specific cleavage of other nucleic acid sequences once the target has been cut (a property known as the "trans-cleavage" activity of Cas proteins).
In this activated trans-cleavage state, the enzyme non-specifically cleaves any surrounding single-stranded nucleic acids. By introducing fluorescently labeled probes into the system, which are then shredded by the enzyme, a detectable signal is generated. This serves as the foundation for various genetic detection technologies.
Taking Cas12a as an example: The activation process of Cas protein trans-cleavage activity
Cas12a processes its own crRNA at the 5' end. Recognition of the dsDNA target begins with PAM recognition by the WED and PI domains, which facilitates the unwinding of the dsDNA target.
The formation of the R-loop is initiated by the binding of the Target Strand (TS) to the crRNA segment. Simultaneous crRNA-TS pairing and TS-Non-Target Strand (NTS) unwinding lead to the formation of a complete R-loop. The hybridization of crRNA-TS DNA induces a conformational change in the REC lobe, resulting in the allosteric uncapping of the catalytic site within the RuvC domain. The NTS binding groove guides the displaced NTS toward the RuvC catalytic site, leading to the cis-cleavage of the NTS.Subsequently, the further unwinding of the PAM-distal TS-NTS duplex allows the TS to enter the RuvC catalytic site, leading to the cis-cleavage of the TS. The PAM-distal dsDNA is released, while the PAM-proximal dsDNA remains bound to the Cas12a-crRNA complex. This locks Cas12a into a catalytically active conformation, allowing for the trans-cleavage of non-target ssDNA.
This can be summarized in the following four steps:
1.The WED and PI domains of the Cas12a protein recognize the PAM sequence of the dsDNA target, facilitating the unwinding of the dsDNA.
2.The crRNA complementarily pairs with the target strand, forming an R-loop with the non-target strand.
3.The REC lobe moves away from the RuvC domain, exposing the RuvC catalytic site, which then performs cis-cleavage of both the non-target and target strands.
4.The PAM-distal dsDNA is released, while the Cas12a-crRNA complex remains bound to the proximal dsDNA, thereby activating the trans-cleavage activity of Cas12a.
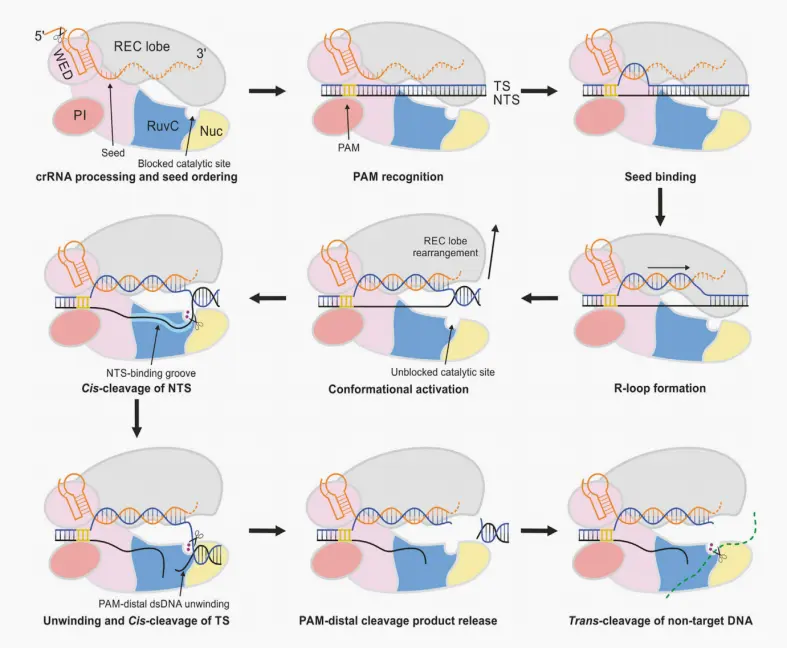
Figure 3. Model of Cas12a-mediated cis- and trans-cleavage of DNA.
In summary, CRISPR detection is a process in which a target nucleic acid activates the trans-cleavage activity of Cas proteins, which then shred fluorescently labeled probes within the system to release a fluorescent signal, thereby completing the detection of the target nucleic acid.