[Quality Share]A Comprehensive Guide to CRISPR Detection
CRISPR Detection

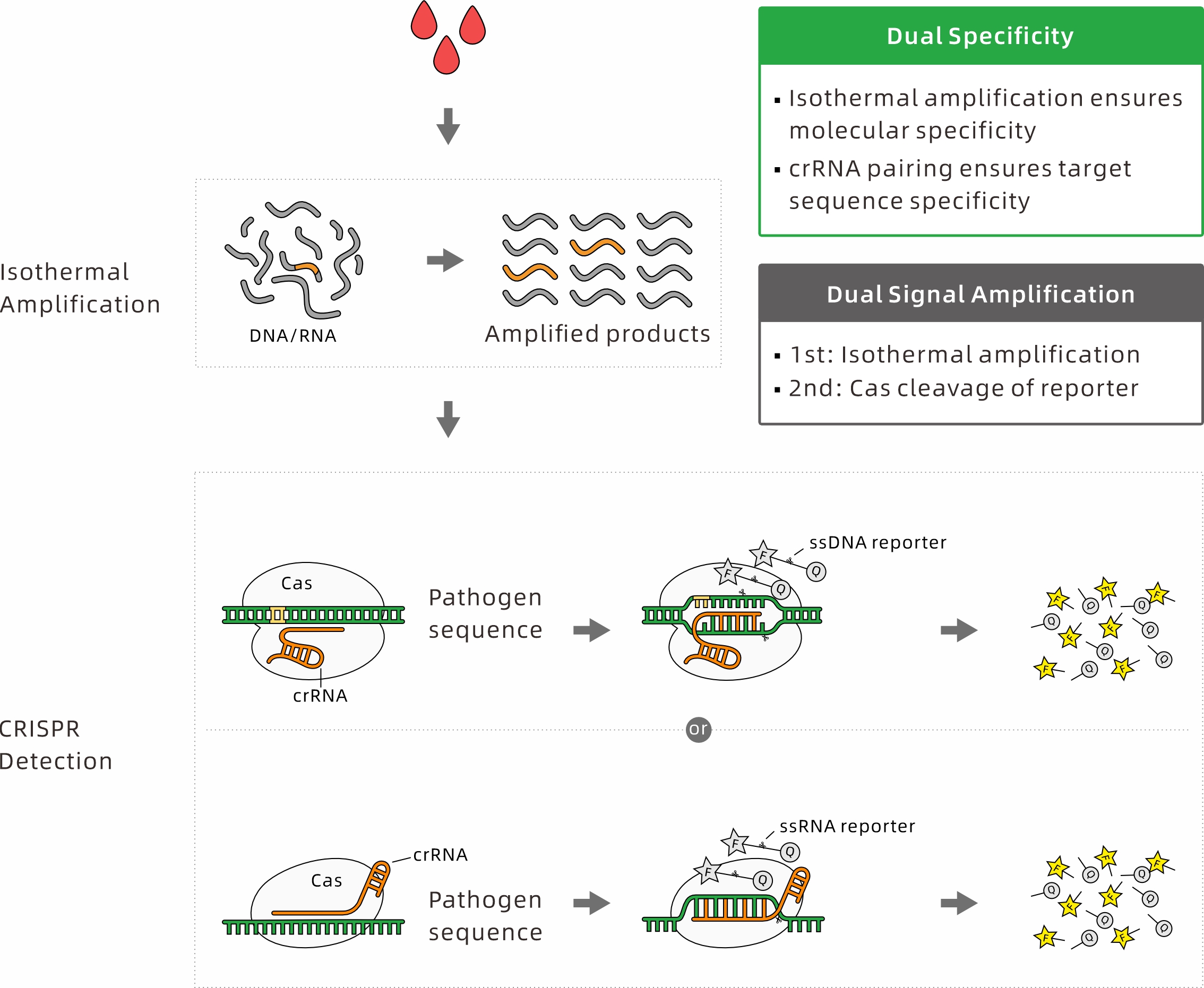
CRISPR detection technology is an analytical approach developed from the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system. In this method, Cas proteins specifically recognize the target nucleic acid sequence, and their trans-cleavage activity subsequently cuts fluorescently labeled reporter probes, generating a measurable fluorescence signal that indicates the presence of the target.
This technique harnesses the unique target recognition and collateral cleavage capabilities of the CRISPR-Cas system to enable rapid, sensitive, and specific detection of DNA or RNA sequences. With its outstanding advantages—including high sensitivity, strong specificity, cost-effectiveness, and versatility—CRISPR-based detection has been widely applied in various fields such as environmental monitoring, animal disease diagnostics, clinical pathogen detection, pet disease screening, and food safety testing.
EDITGENE has compiled some essential knowledge about CRISPR detection, hoping it will be helpful to you.
• Cas12a (Cpf1): A DNA endonuclease that specifically recognizes double-stranded DNA (dsDNA) sequences containing a PAM site (5′-TTTV-3′) and introduces staggered double-strand breaks. Unlike its activity on dsDNA, Cas12a can also cleave single-stranded DNA (ssDNA) targets in a PAM-independent manner.
• Cas12b: A guide RNA-dependent nuclease that recognizes dsDNA with a PAM sequence of 5′-TTV-3′. It generates cohesive double-strand breaks and exhibits higher cleavage activity at elevated temperatures (45-65 °C), making it particularly suitable for coupling with isothermal amplification methods such as LAMP.
• Cas13a: Guided by crRNA, Cas13a targets and cleaves specific single-stranded RNA (ssRNA). Once activated by binding its RNA target, Cas13a displays collateral cleavage activity, indiscriminately cutting nearby ssRNA reporters to generate a detectable signal.
EDITGENE has established an in-house protein expression and purification platform capable of producing Cas12 and Cas13 enzymes with high purity, activity, and recovery efficiency. The detection sensitivity can reach the attomole (amol) level.
|
Cas Protein |
Target Type |
PAM/PFS |
Remarks |
|
LbCas12a |
DNA |
5’TTTV |
|
|
AapCas12b |
DNA |
5’TTV |
Optimal at 40-60 °C, suitable for LAMP coupling |
|
LwaCas13a |
RNA |
PFS-3’A、U、C |
|
|
LbuCas13a |
RNA |
PFS-3’A、U、C |
Produces low background fluorescence |
The crRNA (CRISPR RNA) is a key component of the CRISPR-Cas system. It recognizes and guides the Cas enzyme to the target nucleic acid sequence through complementary base pairing, enabling precise cleavage or editing. The design of crRNA critically influences the sensitivity and specificity of the detection system. Therefore, it is recommended to design and screen multiple crRNAs in vitro to identify the one with optimal performance.
Key considerations for crRNA design and synthesis:
1. Target sequence selection: The choice of the target sequence depends on the functional region of the gene and the specific research purpose. For typing detection, strain- or variant-specific regions should be selected; for broad-spectrum detection, conserved gene regions are preferred.
2. PAM sequence selection: Select the canonical PAM sequence whenever possible; secondary PAM sequences can be used if the canonical site is unavailable.
3.
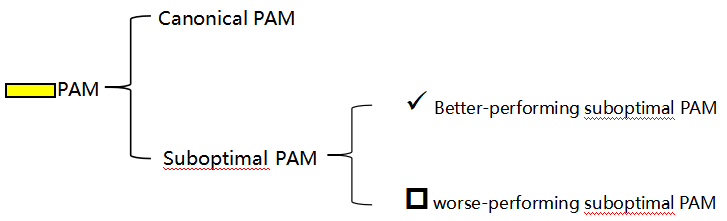
4. Avoiding off-target effects: Ensure that the designed target site does not share significant homology with non-target genes. This can be verified through sequence alignment tools such as BLAST.
5. Minimizing secondary structures: Avoid target regions likely to form stable secondary structures that may interfere with Cas-crRNA binding or cleavage efficiency.
6. crRNA evaluation and validation: Computational tools for sequence alignment and structure prediction can be used to assess crRNA specificity and activity. Final validation should be performed experimentally to confirm the crRNA’s cutting efficiency and target selectivity.
Common crRNA Template Sequences:
The following are commonly used crRNA design templates for various Cas enzymes. In each sequence, N represents the spacer region that is complementary to the specific target sequence.
- • LbCas12a:
5'AAUUUCUACUAAGUGUAGAUNNNNNNNNNNNNNNNNNNNN 3' - • AapCas12b:
5'GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCU
UCUCAAAUCUGAGAAGUGGCACNNNNNNNNNNNNNNNNNNNN 3' - • LwaCas13a:
5'GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACNNNNNNNNNNNNNNNNNNNN 3'(the recommended spacer length is 20-28 nt) - • LbuCas13a:
5'GGACCACCCCAAAAAUGAAGGGGACUAAAACNNNNNNNNNNNNNNNNNNNN 3'
-
III Selection of Isothermal Amplification Systems
Isothermal amplification technology is one of the key methods for achieving rapid and precise nucleic acid detection. Compared with conventional PCR, it offers greater simplicity and faster reaction times, making it especially advantageous for on-site testing or in resource-limited settings. Below are several commonly used isothermal amplification systems:
1. LAMP(Loop-Mediated Isothermal Amplification)
-
LAMP relies on a specially designed set of primers and the strand-displacement activity of Bst DNA polymerase. It operates at a constant temperature (typically 60-65°C) and achieves efficient amplification within a short time (usually less than one hour). The results can often be observed directly by the naked eye, for example, through the addition of fluorescent dyes.
2. RPA(Recombinase Polymerase Amplification)
-
This technique functions at or near room temperature (37-42°C) and does not require specialized equipment. RPA uses recombinase enzymes to unwind double-stranded DNA and can amplify target DNA within a short period, making it highly suitable for rapid detection applications.
3. NASBA(Nucleic Acid Sequence-Based Amplification)
-
The NASBA reaction is performed under isothermal conditions (41-42°C), producing either single-stranded DNA or double-stranded RNA as amplification products. It is highly sensitive for detecting low-abundance RNA targets, though it requires more stable reagents and involves a relatively complex operation process.
When selecting an isothermal amplification system, multiple factors must be considered, including the experimental purpose, target sequence characteristics, experimental conditions, and the stability and efficiency of the amplification system. Two key factors to consider are:
- I. Target Sequence Characteristics: Features such as the length of the target sequence, GC content, and the presence of secondary structures can significantly affect the performance of isothermal amplification. Therefore, it is essential to thoroughly analyze the target sequence and choose an amplification system that is compatible with its characteristics.
-
II. Stability and Efficiency of the Amplification System: The stability and efficiency of an isothermal amplification system are critical considerations. A stable system minimizes experimental variability and ensures reliable results, while a highly efficient system shortens reaction times and improves overall experimental throughput.
As a next-generation molecular diagnostic approach, CRISPR-based detection requires careful selection of the appropriate Cas enzyme, optimized crRNA sequences, and a suitable isothermal amplification system prior to experimentation. These choices have a profound impact on the specificity and sensitivity of the subsequent detection assay.
African Swine Fever Virus (ASFV) Detection
Results:
Using the ASFV 1070 target, the one-tube CRISPR/Cas12a detection assay can detect 100 copies of viral nucleic acid within 10 minutes, and 10-50 copies within 20 minutes.
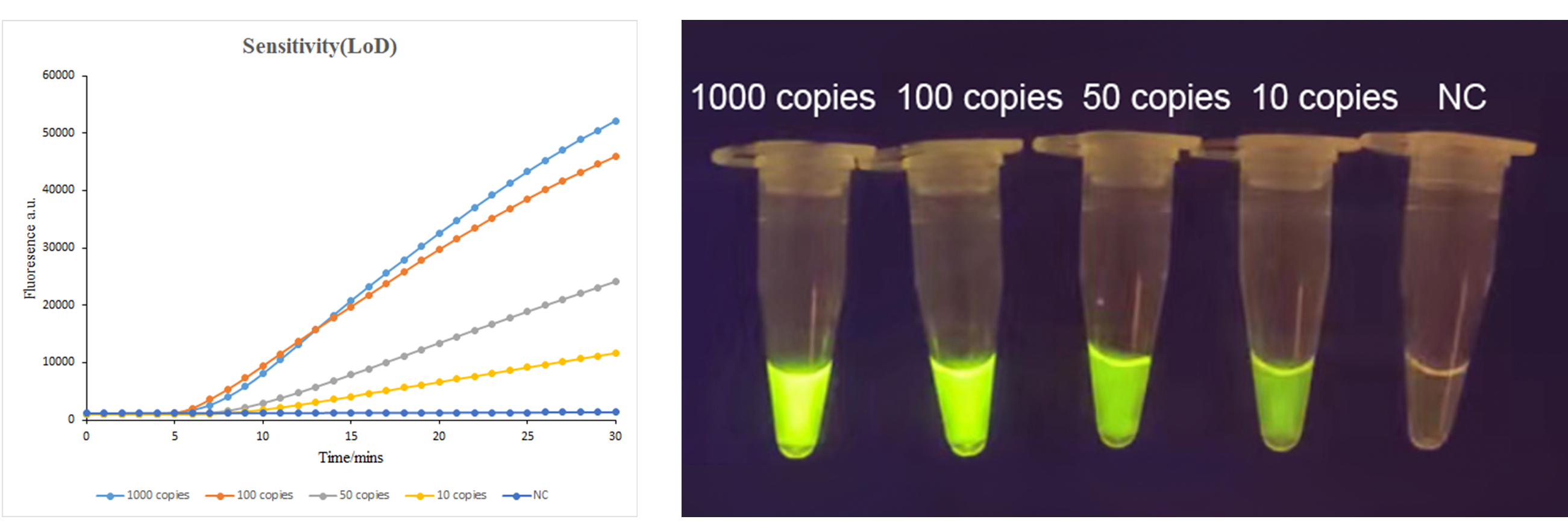









![[Quality Share]A Comprehensive Guide to CRISPR Detection](/uploads/20250328/ESzk5OC49wpxIHVv_3cbfa5e98ea1d238127fe23c72b0f4b2.png)

Comment (4)