[Quality Share] Efficient Generation of CRISPR Point Mutations Using Donor Template Strategies
![]()
CRISPR-Cas9 is a powerful genome editing tool commonly used to introduce point mutations, which involve the substitution of a single nucleotide at a specific genomic locus. Such precise editing typically relies on the Homology-Directed Repair (HDR) pathway and requires a donor template to guide accurate DNA repair.
However, residual donor templates or vectors after mutation introduction can pose several challenges, including random integration, additional cytotoxicity or metabolic burden, and the unintended incorporation of selection markers or other non-target sequences into the genome. Therefore, developing effective strategies for donor template elimination is crucial to mitigate these risks.
Based on recent research, this article summarizes commonly used methods for template or vector removal, including transient delivery, self-eliminating vectors, and marker excision strategies. These approaches maintain high editing efficiency while effectively reducing residual templates, vectors, and associated marker sequences, offering broad applications in cell line development, animal model generation, and gene therapy.
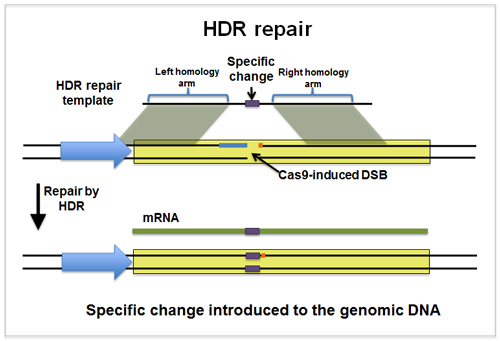
Figure 1. CRISPR-Cas9-mediated HDR repair schematic
01
Fundamental Mechanism of Point Mutation
The CRISPR-Cas9 system employs a single-guide RNA to direct the Cas9 nuclease to a specific genomic locus, generating a DNA double-strand break (DSB). For point mutation editing, the cell can repair the break via the Homology-Directed Repair (HDR) pathway, using a donor template, such as a single-stranded oligonucleotide (ssODN) or a plasmid with homology arms, to precisely replace the target base.
The design of the donor template is critical. The template must carry the desired mutation and include homology arms of appropriate length. Typically, ssODNs contain homology arms of approximately 30-70 base pairs, whereas plasmid donors generally require homology arms of 500 bp or more to achieve higher HDR efficiency.
Studies have shown that the closer the target mutation site is to the sgRNA cleavage site, the higher the editing efficiency.
02
Donor Template Removal Strategies and Their Application
in Efficient Generation of Mutant Cell Lines
in Efficient Generation of Mutant Cell Lines
During HDR-mediated point mutation editing, residual donor templates can pose risks such as random genomic integration, cellular toxicity, or other effects associated with high template concentrations. Therefore, the effective removal of donor templates after the desired mutation is introduced is a critical step to improve cell viability and clonal purity.
Current strategies for template elimination vary depending on the type of donor template (e.g., ssODNs versus plasmid donors) and the host system (e.g., bacterial or mammalian cells). Selecting an appropriate strategy allows for rapid removal of the donor template, thereby facilitating a more efficient and controlled process for generating mutant cell lines.
✅ Transient Template Delivery
✅ Controllable Vector Elimination
Current strategies for template elimination vary depending on the type of donor template (e.g., ssODNs versus plasmid donors) and the host system (e.g., bacterial or mammalian cells). Selecting an appropriate strategy allows for rapid removal of the donor template, thereby facilitating a more efficient and controlled process for generating mutant cell lines.
✅ Transient Template Delivery
① Principle
Non-integrating donor templates, such as ssODNs, are directly introduced into cells via microinjection or electroporation. These templates do not replicate and are naturally diluted or degraded during cell division, eliminating the need for additional removal steps.
Non-integrating donor templates, such as ssODNs, are directly introduced into cells via microinjection or electroporation. These templates do not replicate and are naturally diluted or degraded during cell division, eliminating the need for additional removal steps.
② Application Example
In a rabbit PCSK9 S386A point mutation model, Cas9 mRNA, sgRNA, and ssODN were co-microinjected into zygotes. The donor template exists only transiently within the repair window and naturally disappears afterward, allowing direct generation of homozygous or heterozygous mutant offspring without residual vectors. This approach offers high efficiency and low integration risk in animal model generation.
In a rabbit PCSK9 S386A point mutation model, Cas9 mRNA, sgRNA, and ssODN were co-microinjected into zygotes. The donor template exists only transiently within the repair window and naturally disappears afterward, allowing direct generation of homozygous or heterozygous mutant offspring without residual vectors. This approach offers high efficiency and low integration risk in animal model generation.
③ Advantages
Simple procedure and non-toxic; when combined with RNP complex delivery, HDR efficiencies of 20-86% can be achieved.
Simple procedure and non-toxic; when combined with RNP complex delivery, HDR efficiencies of 20-86% can be achieved.
④ Limitations
Suitable for small-scale mutations; larger templates require optimization.
Suitable for small-scale mutations; larger templates require optimization.
✅ Controllable Vector Elimination
① Principle
Donor templates can be selectively removed using self-eliminating vectors, such as the EXIT genetic circuit. These vectors consist of a control module (including a tightly regulated promoter like P_Bad and a nuclease gene such as I-SceI) and an elimination module (containing an antibiotic resistance gene and recognition sequences). After editing, the addition of an inducer (e.g., L-arabinose) activates the nuclease, which linearizes and degrades the vector, effectively removing the donor template.
Donor templates can be selectively removed using self-eliminating vectors, such as the EXIT genetic circuit. These vectors consist of a control module (including a tightly regulated promoter like P_Bad and a nuclease gene such as I-SceI) and an elimination module (containing an antibiotic resistance gene and recognition sequences). After editing, the addition of an inducer (e.g., L-arabinose) activates the nuclease, which linearizes and degrades the vector, effectively removing the donor template.
② Application Example
Donor templates can be selectively removed using self-eliminating vectors, such as the EXIT genetic circuit. These vectors consist of a control module (including a tightly regulated promoter like P_Bad and a nuclease gene such as I-SceI) and an elimination module (containing an antibiotic resistance gene and recognition sequences). After editing, the addition of an inducer (e.g., L-arabinose) activates the nuclease, which linearizes and degrades the vector, effectively removing the donor template.
Donor templates can be selectively removed using self-eliminating vectors, such as the EXIT genetic circuit. These vectors consist of a control module (including a tightly regulated promoter like P_Bad and a nuclease gene such as I-SceI) and an elimination module (containing an antibiotic resistance gene and recognition sequences). After editing, the addition of an inducer (e.g., L-arabinose) activates the nuclease, which linearizes and degrades the vector, effectively removing the donor template.
③ Advantages
Modular design and one-step operation; reduces metabolic burden and supports iterative editing; more versatile than temperature-sensitive vectors.
Modular design and one-step operation; reduces metabolic burden and supports iterative editing; more versatile than temperature-sensitive vectors.
④ Limitations
Modular design and one-step operation; reduces metabolic burden and supports iterative editing; more versatile than temperature-sensitive vectors.
Modular design and one-step operation; reduces metabolic burden and supports iterative editing; more versatile than temperature-sensitive vectors.
✅ Marker Excision Strategy
① Principle
Donor plasmids not only carry the desired point mutation and homology arms but also include a selectable marker near the mutation site (e.g., PGK-Puro^R) flanked by a pair of loxP sites. Following a Cas9-induced double-strand break (DSB), the HDR pathway integrates the fragment containing both the mutation and the marker into the genome, allowing positive selection of correctly edited cells. Subsequently, the marker cassette can be excised using CRE recombinase, effectively removing both the donor template and the selectable marker.
Donor plasmids not only carry the desired point mutation and homology arms but also include a selectable marker near the mutation site (e.g., PGK-Puro^R) flanked by a pair of loxP sites. Following a Cas9-induced double-strand break (DSB), the HDR pathway integrates the fragment containing both the mutation and the marker into the genome, allowing positive selection of correctly edited cells. Subsequently, the marker cassette can be excised using CRE recombinase, effectively removing both the donor template and the selectable marker.
② Application Example
In patient-derived iPS cells, this approach has been used to correct LRRK2 G2019S or AARS1 R329H point mutations. By targeting adjacent intronic sites, donor integration is highly efficient, and CRE-mediated excision successfully removes the marker without detectable off-target effects. This “low-stress” workflow is suitable for disease model studies and enables rapid generation of isogenic control cell lines.
In patient-derived iPS cells, this approach has been used to correct LRRK2 G2019S or AARS1 R329H point mutations. By targeting adjacent intronic sites, donor integration is highly efficient, and CRE-mediated excision successfully removes the marker without detectable off-target effects. This “low-stress” workflow is suitable for disease model studies and enables rapid generation of isogenic control cell lines.
③ Advantages
Positive selection enriches correctly edited clones, improving efficiency; suitable for mammalian cells.
Positive selection enriches correctly edited clones, improving efficiency; suitable for mammalian cells.
④ Limitations
Requires additional transfection steps; loxP sites may remain in the genome.
Requires additional transfection steps; loxP sites may remain in the genome.
03
Summary
In summary, CRISPR-Cas9 technology, via the HDR pathway, offers a powerful solution for introducing precise point mutations. Efficient and safe elimination of residual donor templates—critical for preventing random integration, cytotoxicity, and other potential risks—remains a key challenge in this process.
The strategies reviewed here, including transient template delivery, controllable vector elimination, and marker excision, provide practical approaches to address this challenge. These methods not only significantly improve clone purity and cell survival but also reduce the long-term burden of exogenous DNA on the cells.
With continued advances in vector design and delivery technologies, donor template removal strategies will become increasingly precise and universally applicable. This will not only accelerate the generation of cell lines and animal models in basic research but also offer safer and more efficient technical support for clinical gene therapy, advancing precision medicine toward a scarless and controllable paradigm.
The strategies reviewed here, including transient template delivery, controllable vector elimination, and marker excision, provide practical approaches to address this challenge. These methods not only significantly improve clone purity and cell survival but also reduce the long-term burden of exogenous DNA on the cells.
With continued advances in vector design and delivery technologies, donor template removal strategies will become increasingly precise and universally applicable. This will not only accelerate the generation of cell lines and animal models in basic research but also offer safer and more efficient technical support for clinical gene therapy, advancing precision medicine toward a scarless and controllable paradigm.
With over a decade of experience in gene editing, EDITGENE has developed the Bingo™ platformbased on cutting-edge Prime Editing (PE) technology. This platform integrates highly efficient sgRNA design with a rigorous monoclonal screening system, enabling the customized generation of various types of precise point mutation cell lines for our customers.
Recent Blogs
1. [Literature Review] Decoding the Latest Nature Paper from David Liu’s Team: How Gene Editing Teaches Cells to “Ignore Errors”
2. [Literature Review] Knocking Out the HO-1 Gene Makes Tumor Cells More
3. [Quality share] MDA-MB-231 Luciferase Stable Cell Line: Empowering Triple-Negative Breast Cancer Metastasis Research
2. [Literature Review] Knocking Out the HO-1 Gene Makes Tumor Cells More
3. [Quality share] MDA-MB-231 Luciferase Stable Cell Line: Empowering Triple-Negative Breast Cancer Metastasis Research












![[Quality Share] Efficient Generation of CRISPR Point Mutations Using Donor Template Strategies](/uploads/20250328/ESzk5OC49wpxIHVv_3cbfa5e98ea1d238127fe23c72b0f4b2.png)

Comment (4)