[Literature Review] Decoding the Latest Nature Paper from David Liu’s Team: How Gene Editing Teaches Cells to “Ignore Errors”
![]()
On November 19 this year, the field of gene editing witnessed another highly anticipated breakthrough. David Liu and colleagues published their latest study in Nature, introducing a novel gene editing strategy—PERT (Prime Editing-mediated Readthrough of Premature Termination Codons)—which offers a potential universal therapeutic approach for a variety of genetic diseases caused by nonsense mutations.

Original link: https://www.nature.com/articles/s41586-025-09732-2
In traditional gene therapy approaches, each mutation typically requires a dedicated editing strategy. This not only increases cost and development time but also limits accessibility for patients with rare diseases. PERT aims to overcome this limitation.
01
Nonsense Mutations and Their Challenges
Nonsense mutations convert codons that normally encode amino acids into stop codons, resulting in prematurely truncated proteins. These mutations account for approximately 24% of all pathogenic variants cataloged in the ClinVar database (a repository of clinically relevant genetic variants and supporting evidence maintained by NCBI), yet they are distributed across many different genes, making it difficult to address them with a single therapeutic tool.
02
Core Principle of PERT
To understand PERT, it helps to first consider how cells synthesize proteins: mRNA serves as the “instruction manual” for proteins, while tRNAs act as the delivery workers, carrying amino acids and translating the instructions into a complete protein. In the manual, there are three specific “stop signals”—UAA, UAG, and UGA—that tell the ribosome to terminate translation. Nonsense mutations introduce these stop signals prematurely, causing proteins to be truncated and lose their normal function.
The research team engineered a “suppressor tRNA” that can read through these erroneous stop codons. When encountering a premature termination signal, this tRNA inserts an amino acid instead of halting translation, allowing the protein to be fully synthesized. Using prime editing, the team then stably integrated this suppressor tRNA into the cellular genome, enabling long-term expression of tRNAs that effectively “ignore” premature stop signals.
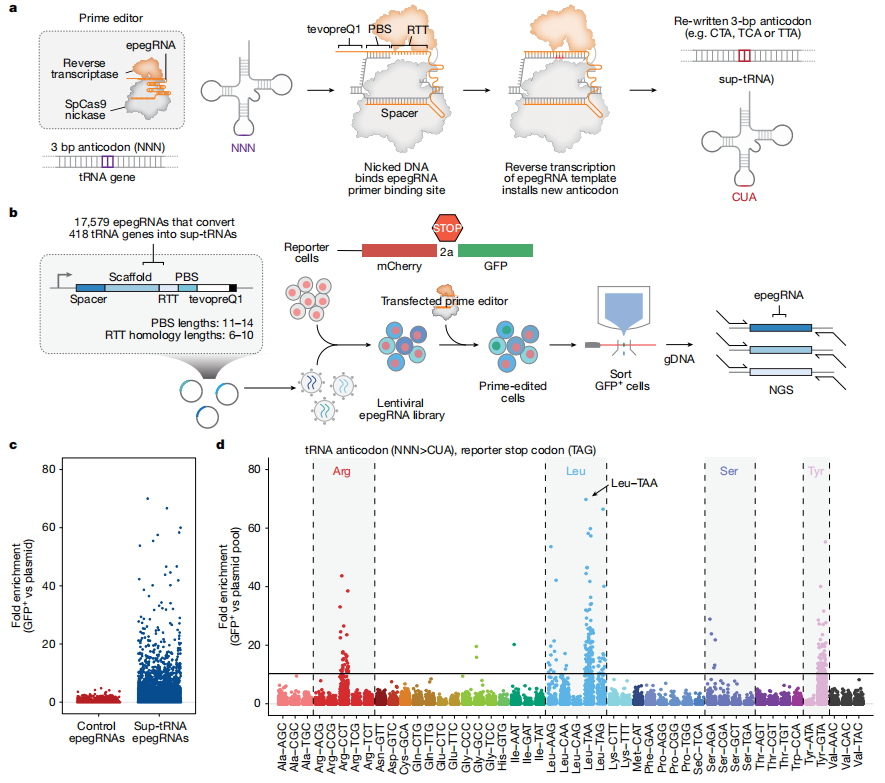
Fig. 1 Prime editing-mediated conversion of endogenous tRNAs to sup-tRNAs in mammalian cells
The core concept of PERT is not to “correct the mutation,” but rather to enable cells to continue synthesizing proteins normally in the presence of a nonsense mutation. Since the common problem across these diseases is the “erroneous stop signal,” PERT simply allows the cell to bypass it.
03
Why PERT Can Address Multiple Diseases
Although the genes involved differ among various diseases, they share a common challenge: the presence of nonsense mutations that prematurely terminate protein synthesis. PERT specifically targets this shared issue, meaning that, in principle, any mutation of this type can be addressed using the same approach, allowing proper protein synthesis.
To identify the optimal sup-tRNA variants, the research team performed saturated mutagenesis of tRNA genes, evaluating multiple combinations accessible via Prime Editing. By constructing various mutations in the anticodon loop and related regions, they screened for engineered sup-tRNAs with high readthrough efficiency. These variants achieved protein yields exceeding 35% relative to wild-type, providing a foundational toolkit for addressing diverse nonsense mutations.
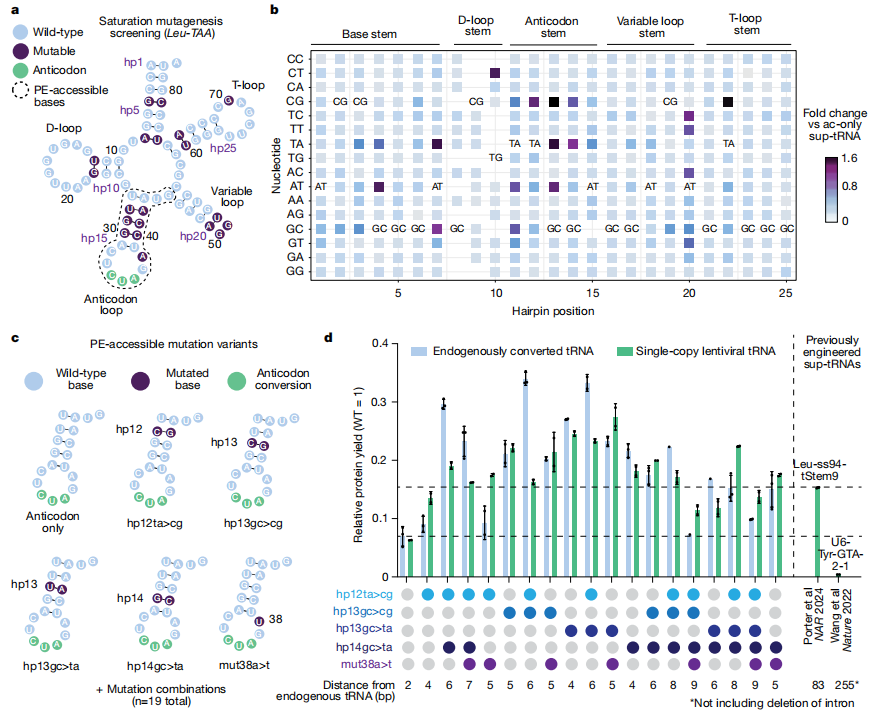
Fig. 2 Identifying prime editing-accessible sup-tRNA mutation combinations
04
PERT Optimization and Cellular Validation
To further optimize the Prime Editing system, the research team designed multiple Prime Editor variants and epegRNA architectures, testing editing efficiencies across different combinations of PBS (primer binding site) and RTT (reverse transcription template) lengths. They also evaluated the impact of MLH1dn (MutL homolog 1 dominant negative, a DNA repair inhibitor) on editing outcomes. This systematic optimization enabled efficient installation of sup-tRNAs, laying the groundwork for subsequent functional validation in disease models.
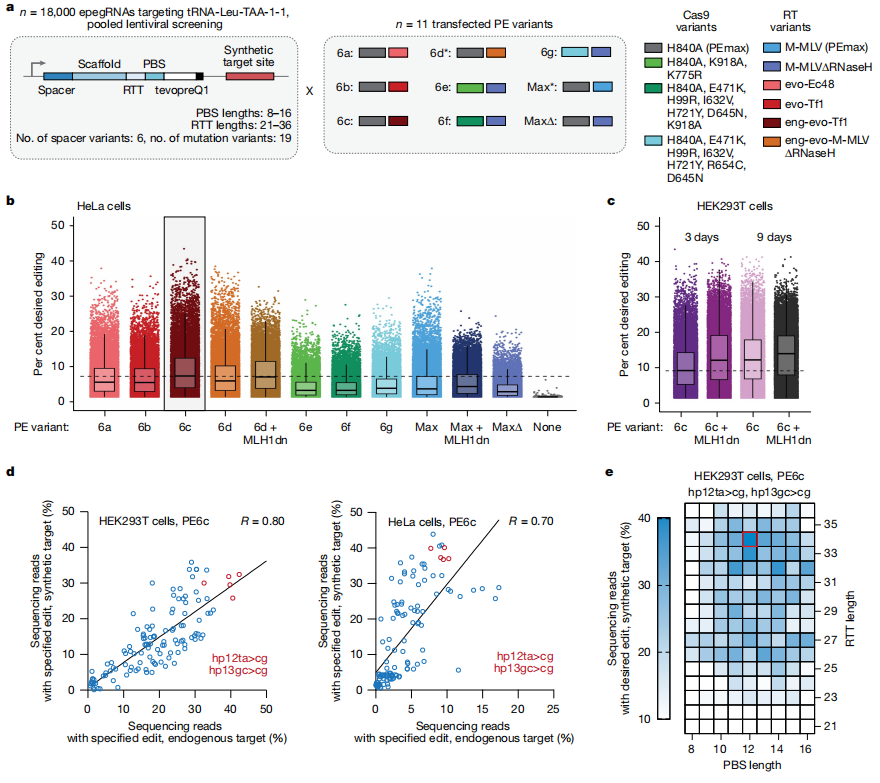
Fig. 3 Optimization of a prime editing strategy for engineered sup-tRNA installation
Following sup-tRNA installation, protein or enzymatic activity was restored to 17%-70%, with partial recovery of full-length protein observed in some NPC1 models. Most mRNAs remained stable at the nonsense mutation sites, indicating that PERT effectively overcomes NMD (Nonsense-Mediated Decay) and preserves transcripts.To evaluate PERT’s practical efficacy, the team tested it across several genetic disease cell models harboring nonsense mutations, including Batten disease, Tay-Sachs disease, and Niemann-Pick type C1. In six HEK293T disease models, installation of engineered sup-tRNAs achieved editing efficiencies of 56-84%, ultimately restoring TPP1, HEXA, and NPC1 protein or enzymatic activity—reaching or even exceeding known therapeutic thresholds.
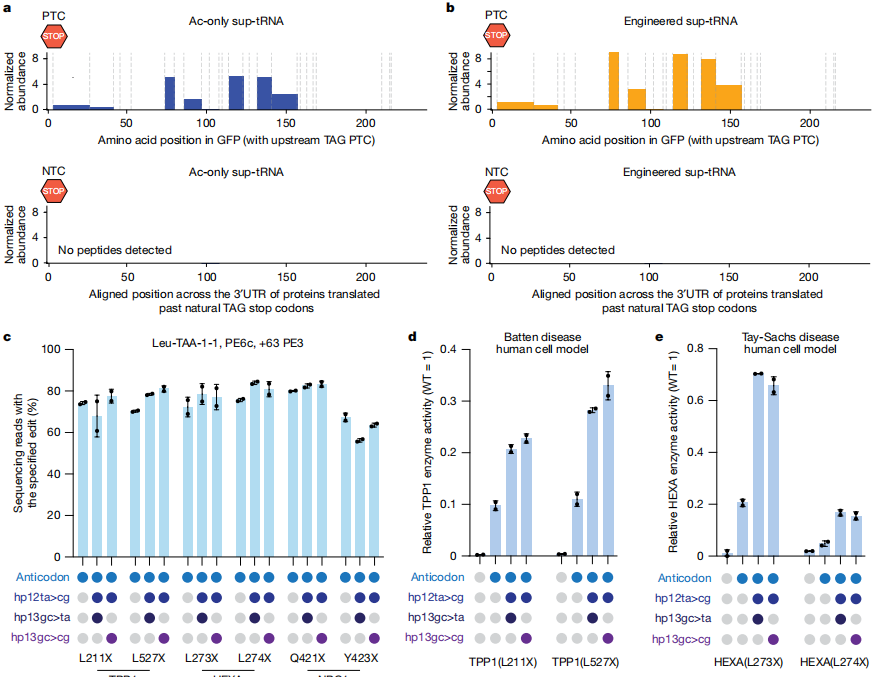
Fig. 4 Prime editing-installed sup-tRNAs can rescue protein expression across diverse disease contexts
05
In Vivo Validation of PERT
In in vivo experiments, the team delivered the Prime Editing system along with sup-tRNA into the brains of neonatal mice using AAV9. Readthrough efficiency was assessed via a GFP reporter system, with edited cells exhibiting relative GFP protein levels of 24%-26%, demonstrating that installed sup-tRNAs can generate functional protein in vivo.
Subsequently, in a Hurler syndrome (MPS-I, Idua^W392X mouse model) study, PERT restored approximately 6% of IDUA (α-L-iduronidase, a lysosomal enzyme) activity in affected tissues, with pathological manifestations nearly eliminated. Histological analyses of the brain, liver, spleen, and heart showed that foam cell accumulation, Purkinje cell vacuolization, and glycosaminoglycan (GAG) deposition observed in untreated mice were significantly alleviated or absent in the PERT-treated group, indicating that partial restoration of protein function can yield substantial therapeutic benefits.
Subsequently, in a Hurler syndrome (MPS-I, Idua^W392X mouse model) study, PERT restored approximately 6% of IDUA (α-L-iduronidase, a lysosomal enzyme) activity in affected tissues, with pathological manifestations nearly eliminated. Histological analyses of the brain, liver, spleen, and heart showed that foam cell accumulation, Purkinje cell vacuolization, and glycosaminoglycan (GAG) deposition observed in untreated mice were significantly alleviated or absent in the PERT-treated group, indicating that partial restoration of protein function can yield substantial therapeutic benefits.

Fig. 5 Prime editing generates functional sup-tRNAs to rescue animal models of disease
A comprehensive overview indicates that PERT, by converting endogenous tRNA genes into sup-tRNAs, establishes an editing strategy applicable across multiple nonsense mutation-related diseases. Combined with the durable, single-installation feature of Prime Editing, this approach demonstrates cross-disease potential, effectively embodying a “one intervention, multiple benefits” paradigm.
Both cellular and animal model studies showed that PERT can restore functional protein or enzymatic activity without observable toxicity or off-target effects, laying a foundation for future clinical applications. Looking ahead, PERT could be paired with tissue-specific viral or nanoparticle delivery systems for organ-targeted therapy, or combined with small-molecule drugs to further enhance readthrough efficiency, offering new therapeutic hope for a wider spectrum of genetic disorder patients.
Both cellular and animal model studies showed that PERT can restore functional protein or enzymatic activity without observable toxicity or off-target effects, laying a foundation for future clinical applications. Looking ahead, PERT could be paired with tissue-specific viral or nanoparticle delivery systems for organ-targeted therapy, or combined with small-molecule drugs to further enhance readthrough efficiency, offering new therapeutic hope for a wider spectrum of genetic disorder patients.
With over a decade of experience in gene editing, EDITGENE has developed the Bingo™ platform based on cutting-edge Prime Editing (PE) technology This platform integrates highly efficient sgRNA design with a rigorous monoclonal screening system, enabling the customized generation of various types of precise point mutation cell lines for our customers.

















