[Literature Review] CRISPR/Cas12a Powers a Novel SNP Detection Breakthrough
— The Asymmetric Hybridization-Driven CRISPR/Cas Adapter
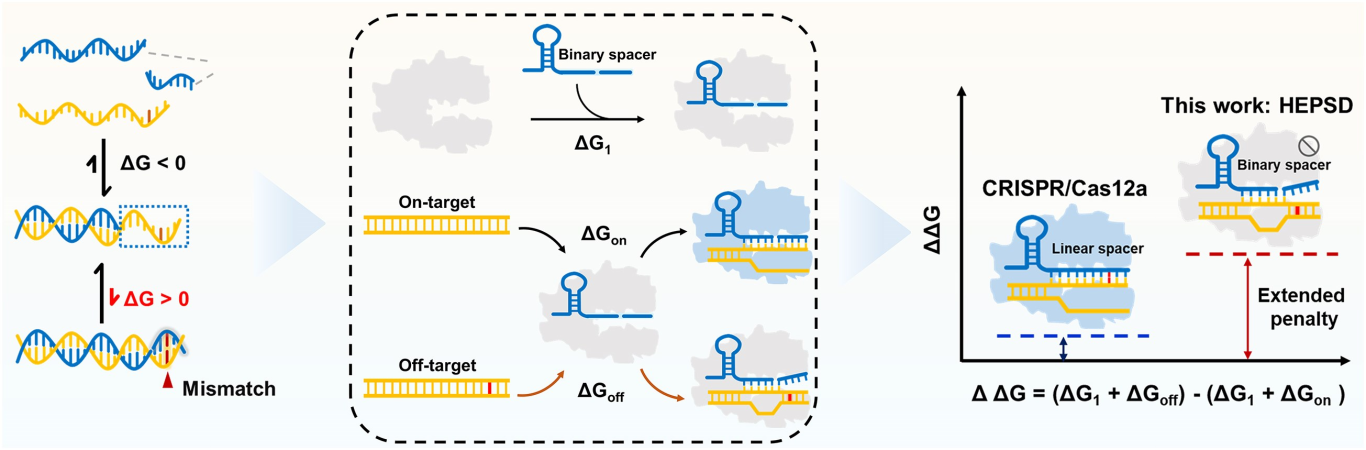
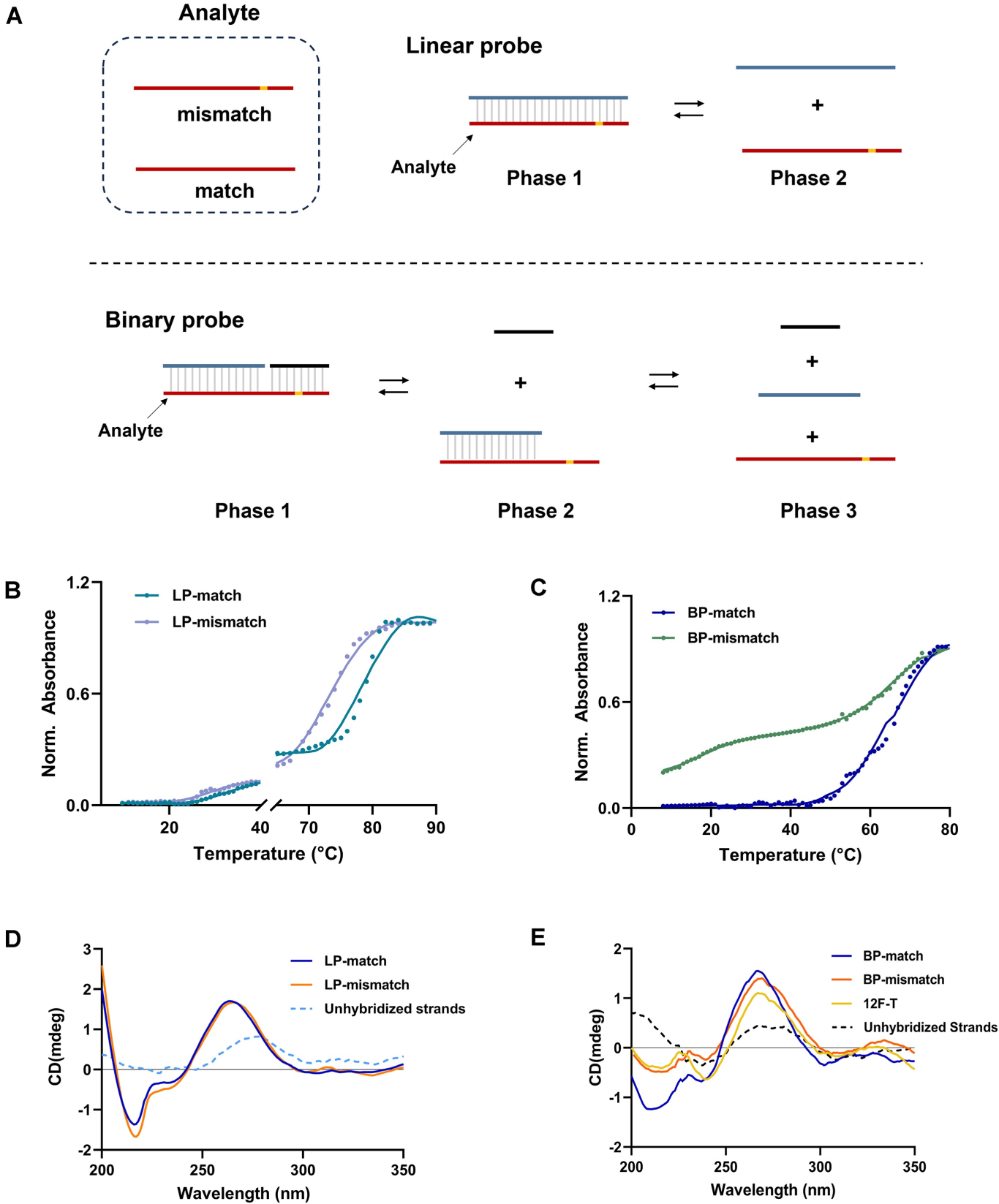
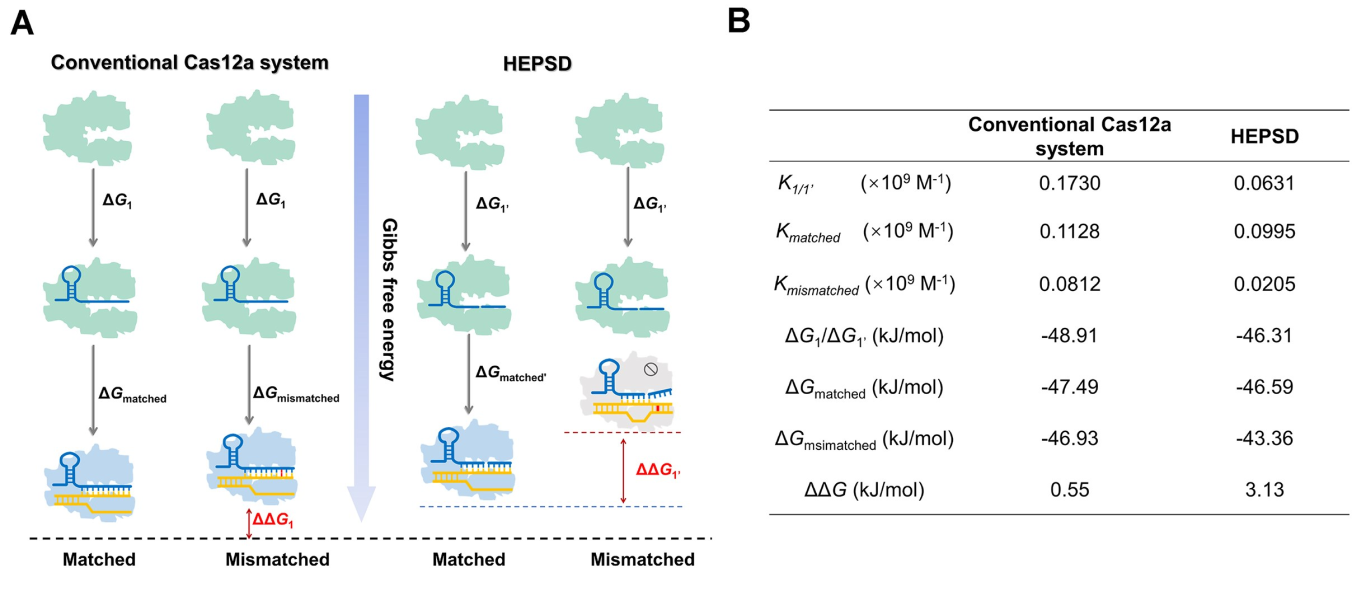
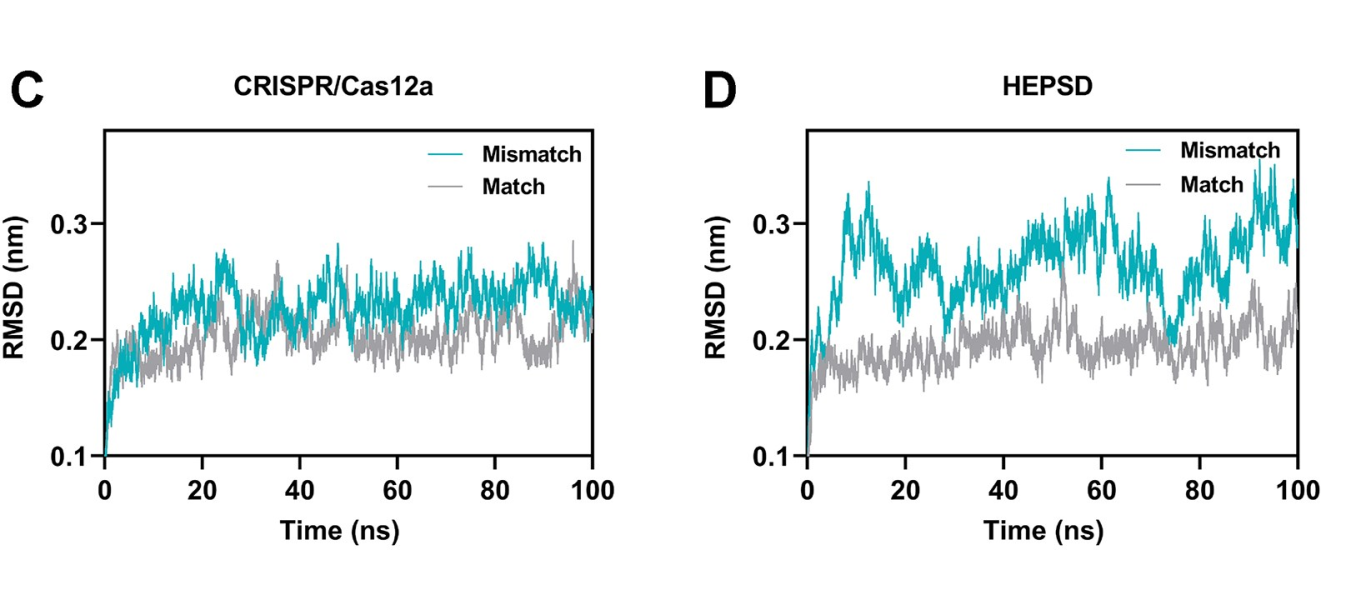
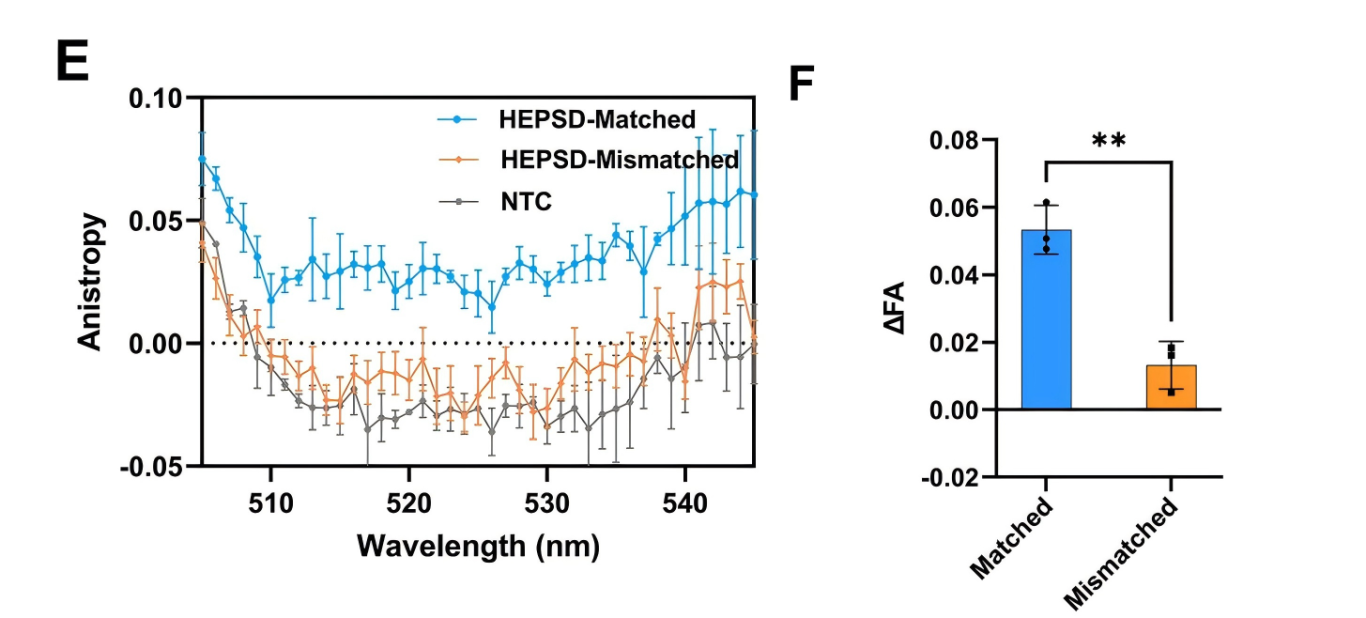
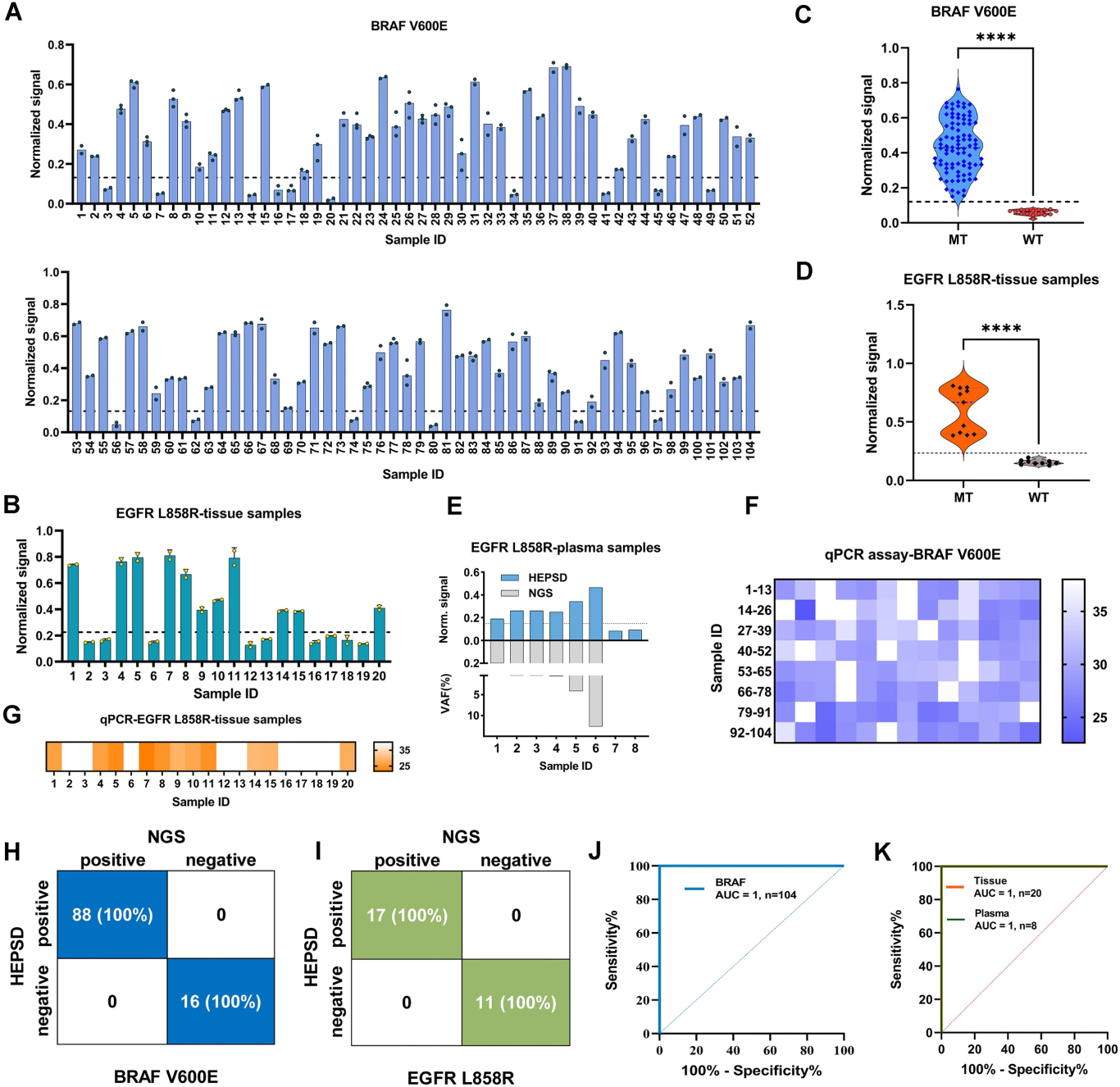
Single-nucleotide variants (SNVs) represent a prevalent form of genetic variation and are known to play critical roles in the pathogenesis of various diseases, including cancers, monogenic disorders, and infectious diseases. The ability to accurately identify SNVs is essential for early diagnosis and the advancement of personalized medicine. However, conventional CRISPR-Cas12a systems face inherent limitations in SNV discrimination, largely due to minimal free energy differences—especially in regions with wobble base pairs or high GC content. To address this limitation, Chinese researchers have developed a novel CRISPR-Cas12a-based detection strategy: the High-Energy Penalty SNV Detection (HEPSD) platform. By incorporating a nonequilibrium hybridization mechanism, the platform effectively enhances the CRISPR system’s ability to detect complex SNV types with greater precision. This study titled “Nonequilibrium hybridization-driven CRISPR/Cas adapter with extended energetic penalty for discrimination of single-nucleotide variants” was published in Nucleic Acids Research (IF: 16.6). Original link: https://doi.org/10.1093/nar/gkaf287 Spotlight 1.The researchers have developed the High-Energy Penalty SNV Detection (HEPSD) platform, leveraging a nonequilibrium hybridization mechanism to substantially enhance the detection of single-nucleotide variants (SNVs). 2.The HEPSD system effectively discriminates challenging SNVs, such as those in high-GC content regions and wobble base pairs, overcoming the limitations of conventional detection methods. 3.Exhibiting remarkable sensitivity, the platform can detect mutations with allele frequencies as low as 0.01%, enabling reliable identification of low-abundance variants. 4.Molecular dynamics simulations combined with fluorescence anisotropy measurements revealed the underlying nonequilibrium hybridization mechanism of HEPSD, offering new insights into the specific regulation of CRISPR systems. Previous studies have shown that traditional CRISPR-Cas12a systems exhibit only minor free energy changes (ΔG) when recognizing mismatched targets, making it difficult to effectively distinguish between wild-type and mutant sequences. To improve sensitivity in detecting SNVs, there is a need for a system that significantly increases the energetic penalty caused by mismatches. To address this, the researches developed a dual-component crRNA architecture that leverages a nonequilibrium hybridization-driven regulatory mechanism to amplify the energetic penalty associated with single-nucleotide mismatches. This design forms the basis of the High-Energetic-Penalty SNV Detection (HEPSD) platform. The platform enables the CRISPR-Cas12a system to preferentially recognize mutations across the entire target region of genomic DNA, while effectively preventing false activation induced by single nucleotide mismatches. Figure 1: Schematic of the HEPSD platform design 1. Thermodynamic Analysis Ultraviolet melting experiments demonstrated that binary probes (BPs) exhibit stable hybridization characteristics across a temperature range of 25°C to 50°C, whereas linear probes (LPs) only show significant hybridization above 68°C. Circular dichroism analysis further revealed that BPs form typical RNA/DNA duplex structures when bound to perfectly matched targets, while the signal markedly decreases in the presence of mismatched targets. These findings indicate that BPs maintain stable hybridization over a broad temperature range and impose a substantial energetic penalty on mismatched targets, thereby enhancing the discrimination of single-nucleotide variants. Figure 2: Characterization of discrimination between LP and BP 2.Fluorescence Quenching Analysis Fluorescence quenching experiments monitoring the binding of Cas12a to RNA (either conventional crRNA or crRNA assisted by binary probes) revealed that the free energy change (ΔG) for the traditional CRISPR-Cas12a system is 0.55 kJ/mol, while the HEPSD platform exhibits a substantially increased ΔG of 3.13 kJ/mol. This indicates that the HEPSD platform imposes a higher energetic barrier when recognizing mismatched targets, effectively suppressing Cas12a activation and thereby improving the discrimination of single-nucleotide variants. Figure 3: Comparative analysis of ΔG and ΔΔG values between conventional CRISPR/Cas12a system and HEPSD platform for matched and mismatched targets 3.Molecular Dynamics Study The results of molecular dynamics study revealed that the RMSD values for matched targets stabilized after 10 ns, while those for mismatched targets exhibited continuous fluctuations. This suggests that the HEPSD platform displays dynamic instability when recognizing mismatched targets but maintains stable binding with matched targets, further supporting its high specificity in detection. Figure 4: RMSD analysis from molecular dynamics simulations of CRISPR/Cas12a (C) and HEPSD (D) with matched and mismatched targets 4.Fluorescence Anisotropy Measurement Fluorescence anisotropy analysis of the HEPSD platform revealed higher anisotropy values when binding to matched targets and significantly lower values for mismatched targets. This indicates weaker binding to mismatched targets and further confirms the platform’s excellent specificity. Figure 5: Fluorescence anisotropy analysis of binding stability between HEPSD and matched or mismatched targets The researches applied the HEPSD platform to test 132 clinical samples covering BRAF V600E and EGFR L858R mutations. Among 104 BRAF V600E samples, the results showed a high concordance with NGS for 88 mutant and 16 wild-type samples. For 20 EGFR L858R samples, 11 mutant and 9 wild-type samples also matched NGS findings. Additionally, in 8 clinical plasma samples, mutation detection by HEPSD was consistent with NGS results. Overall, the HEPSD platform achieved 100% accuracy in clinical sample testing. Figure 6: Clinical analysis of BRAF V600E and EGFR L858R samples In conclusion, the researchers ingeniously redesigned the crRNA hybridization region within the CRISPR/Cas12a system, leveraging a nonequilibrium hybridization-driven mechanism to markedly increase the energetic penalty for single-nucleotide mismatches. Building on this, they successfully developed the high-energetic-penalty single-nucleotide variant detection (HEPSD) platform, significantly enhancing sensitivity for detecting single-nucleotide variants (SNVs). This platform can detect allele frequencies of BRAF V600E and EGFR L858R tumor mutations down to 0.01% and accurately distinguished 132 clinical samples, demonstrating its great potential for applications in clinical molecular diagnostics.


Search
Search
Recent Post
-
![[Literature Review] CRISPR/Cas12a Powers a Novel SNP Detection Breakthrough <br/>— The Asymmetric Hybridization-Driven CRISPR/Cas Adapter](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)
9 June.2025
[Literature Review] CRISPR/Cas12a Powers a Novel SNP Detection Breakthrough
— The Asymmetric Hybridization-Driven CRISPR/Cas Adapter -
![[Client Publication] EDITGENE's Point Mutation Cells Illuminate the Role of TTN Mutations in DNA Repair and Immune Infiltration](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)
4 June.2025
[Client Publication] EDITGENE's Point Mutation Cells Illuminate the Role of TTN Mutations in DNA Repair and Immune Infiltration
-
![[Literature Review] CRISPR screening system reveals synthetic lethality in DNA damage response](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)
27 May.2025
[Literature Review] CRISPR screening system reveals synthetic lethality in DNA damage response
















Comment (4)