[Literature Review] CRISPR Screening Reveals the Critical Role of NUS1 in Prostate Cancer Survival and Growth

One of the key features of prostate cancer is lipid metabolism dysregulation, however, the specific regulatory mechanisms involved in tumorigenesis remain unclear. In April 2025, a research team from the University of Melbourne, Peter MacCallum Cancer Centre, and Monash University published a study in Cell Reports (Q1 TOP, IF: 7.5) titled "Lipid-metabolism-focused CRISPR screens identify enzymes of the mevalonate pathway as essential for prostate cancer growth." The team designed a custom lipid metabolism CRISPR knockout library and identified essential genes for prostate cancer survival. For the first time, they proposed the polyprenol-N-glycosylation axis as a potential therapeutic target.

I. CRISPR-Cas9 Screening Identifies Essential Genes Required for Prostate Cancer Survival
The researchers designed a lipid metabolism-targeted CRISPR-Cas9 knockout library containing 13,500 sgRNAs targeting 1,530 lipid metabolism-related genes. Screening was performed in three prostate cancer cell lines: hormone-sensitive (LNCaP), castration-resistant (C4-2B), and enzalutamide-resistant (MR-49F). A total of 63 common dependency genes with a false discovery rate (FDR) < 5% were identified. These genes were mainly enriched in the mevalonate pathway, including HMGCS1, MVK, and MVD, and the polyprenol-N-glycosylation pathway, including NUS1, DHDDS, DOLK, and DPAGT1.
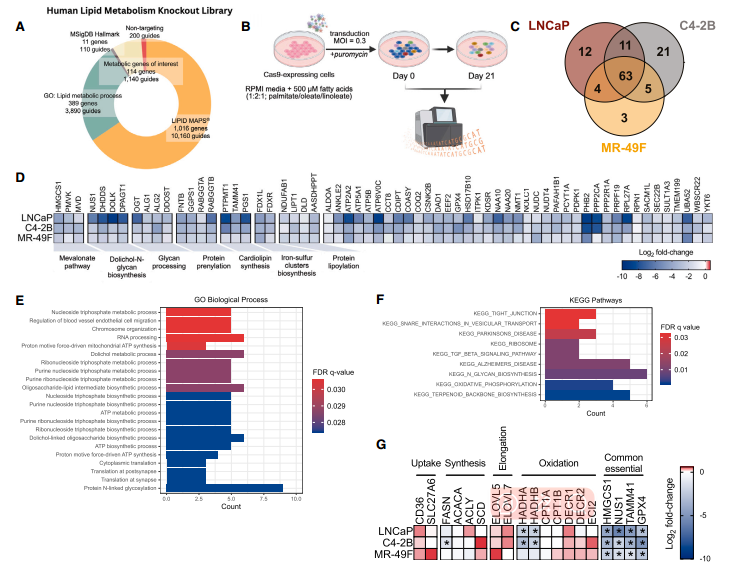
Figure 1. CRISPR-Cas9 KO screening identifies genes essential for prostate cancer survival
II. NUS1 Is a Critical Gene Required for Prostate Cancer Survival
Due to the limited research on the polyprenol-N-glycosylation pathway in prostate cancer, the researchers selected the three most enriched genes from this pathway for further investigation. In C4-2B-Cas9 cells, knockout of NUS1 led to a significant reduction in cell proliferation and an increase in cell death. In an in vivo xenograft model, loss of NUS1 markedly suppressed tumor growth and significantly prolonged mouse survival, whereas knockout of DOLK and DPAGT1 did not show similar effects. These findings indicate that NUS1 plays a vital role in prostate cancer survival.
NUS1 encodes a subunit of cis-prenyltransferase (cis-PTase), which catalyzes the formation of polyprenyl pyrophosphate (Pol-PP) from farnesyl pyrophosphate (FPP)—a key step in protein N-glycosylation.
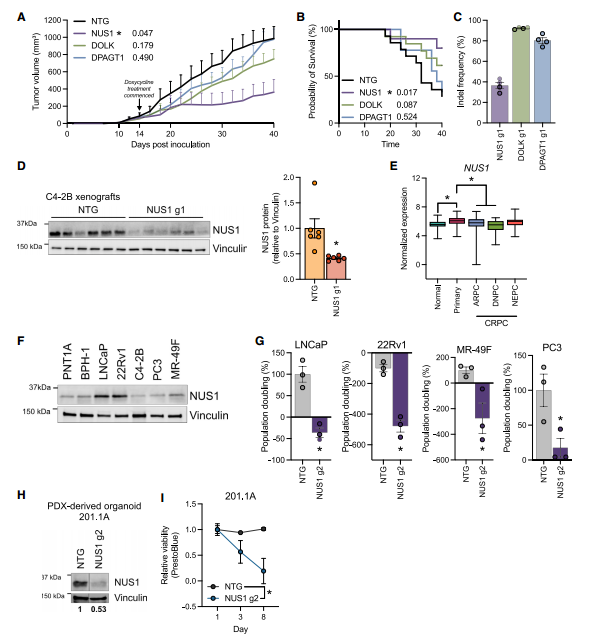
Figure 2. NUS1 is an essential gene for prostate cancer growth
III. Loss of NUS1 Leads to Reduced Cell Viability Associated with ER Stress and G1 Cell Cycle Arrest
① NUS1 Knockout Induces Endoplasmic Reticulum (ER) Stress:
The researchers assessed changes in mRNA and protein expression of ER stress markers (BIP, sXBP-1, CHOP) after NUS1 knockout using RT-qPCR and Western blot. They also analyzed BIP and CHOP protein levels in NUS1 knockout xenograft tumor tissues. The results showed that NUS1 knockout significantly upregulated the mRNA and protein levels of BIP (p < 0.05), sXBP-1, and CHOP. In the xenograft tumors, the protein levels of BIP and CHOP increased 2.1 and 1.8 times, respectively.
② Cell Cycle Arrest at G1 Phase:
Flow cytometry analysis was used to examine the cell cycle distribution of NUS1 knockout cells, and Western blot was performed to measure the phosphorylation levels of p38 MAPK and ERK1/2. The results revealed that NUS1 knockout increased the proportion of cells in G1 phase from 55% to 72%, while the S phase decreased by 40%. The phosphorylation levels of p38 were increased 2-fold, and ERK1/2 phosphorylation levels were upregulated by 1.5 times. These findings suggest that NUS1 loss induces G1 phase arrest and inhibits cell proliferation by activating the p38 MAPK pathway.
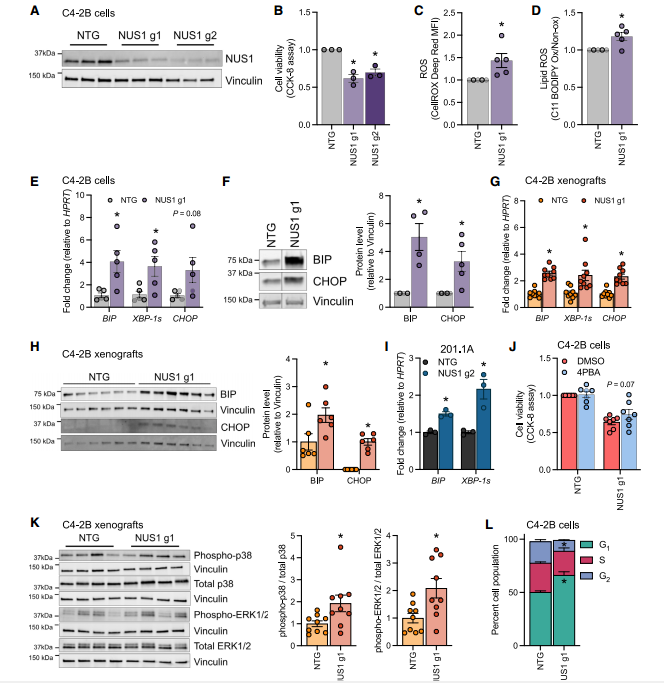
Figure 3. Loss of NUS1 in prostate cancer promotes ER stress and activates p38 and ERK1/2 MAPK
IV. Potential Impact of NUS1 Loss on Treatment
Enzalutamide, which targets androgen receptor (AR) signaling, is currently the first-line treatment for prostate cancer. The researchers evaluated the synergistic effect of enzalutamide by co-administering it with NUS1 knockout in C4-2B organoids. They also analyzed the expression of AR and its target genes (KLK3, NKX3-1) using Western blot and RT-qPCR. The results showed that NUS1 knockout reduced AR protein levels by 40% (p < 0.05) and decreased KLK3 and NKX3-1 mRNA expression by 50% and 60%, respectively. Co-treatment with enzalutamide further reduced the viability of NUS1 knockout organoids by 1.6-fold (p < 0.05). These findings suggest that loss of NUS1 can enhance the effectiveness of anti-androgen therapy.
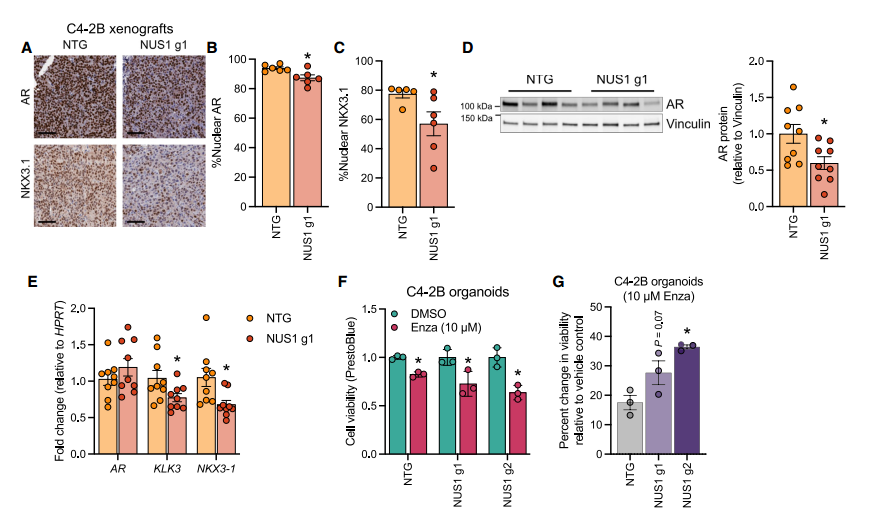
Figure 4. Loss of NUS1 inhibits AR signaling in prostate cancer
In conclusion, this study uses CRISPR screening to reveal for the first time that NUS1 maintains prostate cancer cell survival and AR signaling activity by regulating the polyprenol-N-glycosylation biosynthesis axis. Its loss induces ER stress, G1 phase arrest, and enhances the efficacy of anti-androgen therapy, providing a new insight for targeted treatment of prostate cancer.
EDITGENE specializes in customized gene knockout cell line development, delivering powerful tools that help researchers unlock critical mechanisms and drive discoveries from bench to bedside.Ready to accelerate your gene editing research?Contact us today to tailor a solution for your study.EDITGENE– making target validation more rapid and more precise.
Recent Blogs
- 1. [Literature Review]EDITGENE Supports Research Team in Uncovering New Mechanism of Macrophage Polarization Regulated by Dental Pulp Stem Cells
- 2. [Quality Share]Cracking the Delivery Puzzle: How Can Gene Editing Precisely Reach Target Cells?
- 3. [Literature Review] CRISPR–Csm: Real-time Single-Molecule Tracking of RNA Dynamics in Live Cells
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com
Search
Search
Recent Post
-

29 April.2025
【Literature Review】EZH2 Knockout Mouse Model Sheds Light on Ferroptosis Mechanisms in Pulpitis
-
![[Literature Review] CRISPR Screening Reveals the Critical Role of NUS1 in Prostate Cancer Survival and Growth](/uploads/20250429/st8wmjq4plYCdzgA_53c82bdd67704fe0e159246934f924ee.png)
29 April.2025
[Literature Review] CRISPR Screening Reveals the Critical Role of NUS1 in Prostate Cancer Survival and Growth
-

29 April.2025
EDITGENE Supports Research Team in Uncovering New Mechanism of Macrophage Polarization Regulated by Dental Pulp Stem Cells
















Comment (4)