EDITGENE Supports Research Team in Uncovering New Mechanism of Macrophage Polarization Regulated by Dental Pulp Stem Cells

Background:
Macrophages are key players in immune responses, capable of polarizing into either pro-inflammatory (M1) or anti-inflammatory (M2) phenotypes. M1 macrophages amplify inflammation by releasing pro-inflammatory cytokines, whereas M2 macrophages suppress inflammation and promote tissue repair. Maintaining the balance between these two states is critical for the treatment of inflammatory diseases.
Although mesenchymal stem cells (MSCs) have shown promise in immune modulation, challenges such as limited sources and unclear mechanisms remain. In this study, researchers used conditioned medium derived from dental pulp stem cells (DFMSC-CM) to explore its role in regulating macrophage polarization. The findings shed light on the underlying molecular mechanisms and provide a scientific foundation for developing novel anti-inflammatory therapies.
Abstract:
Macrophages are the “double-edged guardians” of the immune system, and precisely regulating their polarization is a major challenge in treating inflammatory diseases. A research team from Southern Medical University recently published a study in Stem Cells International titled “Conditioned Medium Derived From Human Dental Follicle Mesenchymal Stem Cells Alleviates Macrophage Proinflammatory Responses Through MAPK-ERK-EGR1 Axis.” The study reveals that conditioned medium from dental follicle mesenchymal stem cells (DFMSC-CM) modulates macrophage polarization via the MAPK-ERK-EGR1 signaling axis, offering new insights for anti-inflammatory therapy.
At the heart of this research lies a key experimental tool—the EGR1 knockout THP-1 cell line, developed by EDITGENE, which played a critical role in validating the findings.

I. Cell Isolation and Characterization
The researchers isolated dental pulp stem cells (DFMSCs) from third molar pulp tissue donated by clinical volunteers. These cells were confirmed to exhibit the classical characteristics of mesenchymal stem cells (MSCs), including high expression of surface markers CD90, CD105, and CD73, low expression of CD45 and CD34, and the capacity to differentiate into adipogenic, osteogenic, and chondrogenic lineages.
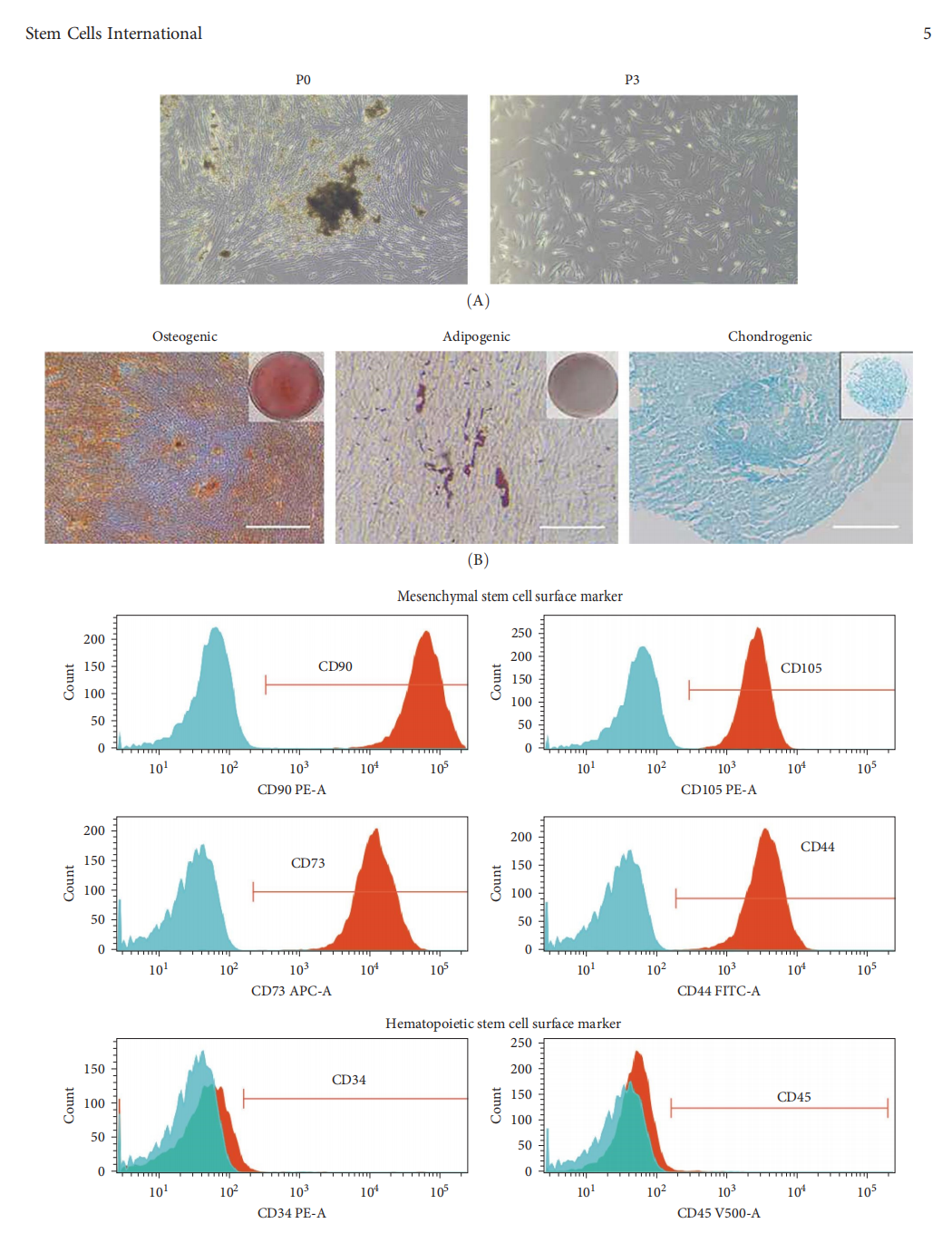
Figure 1: Isolation and characterization of DFMSCs
II. Preparation, Quality Control, and Functional Testing of DFMSC-CM
DFMSC-CM was prepared by collecting serum-free conditioned medium from dental pulp stem cells (DFMSCs), followed by ultrafiltration concentration. Experimental results showed that a protein concentration of 0.02 mg/mL in the CM yielded optimal macrophage viability and the most favorable inflammatory cytokine expression profile. These findings indicate that the preparation process of DFMSC-CM is both stable and reliable.
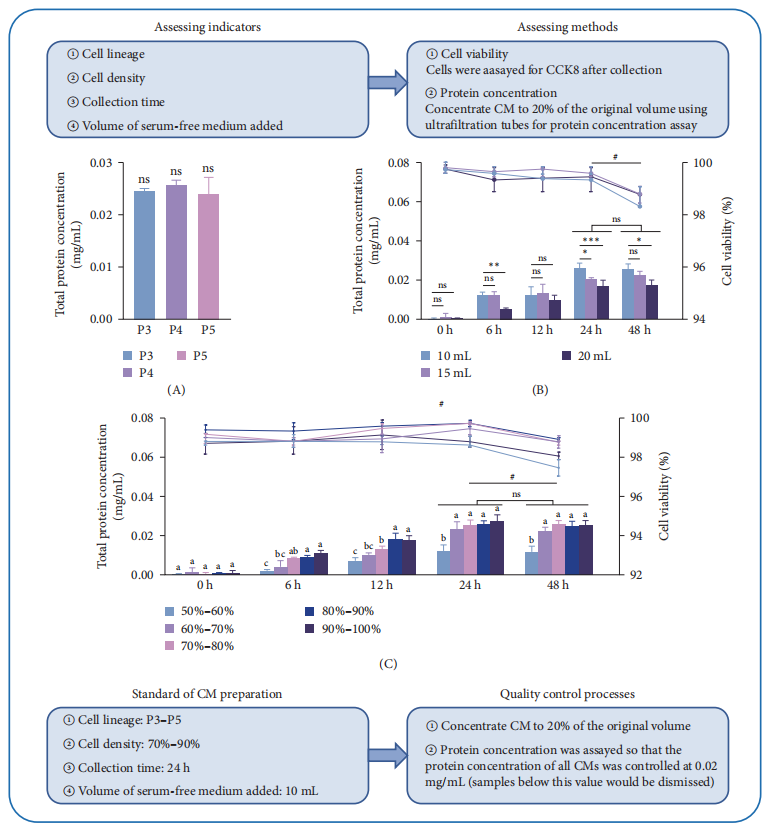
Figure 2: Optimization of DFMSC-CM preparation based on protein concentration and cell viability under different conditions
III. Macrophage Induction and Co-culture
The researchers differentiated THP-1 cells into macrophages and co-cultured them with DFMSC-CM under inflammatory conditions induced by LPS and IFN-γ. The results showed that DFMSC-CM significantly reduced the expression of pro-inflammatory cytokines such as IL-1β and TNF-α, while enhancing the expression of anti-inflammatory cytokines like IL-10 and CCL18. Moreover, DFMSC-CM promoted a shift in macrophage phenotype from pro-inflammatory M1 to anti-inflammatory M2, as evidenced by decreased expression of the M1 marker CD80 and increased expression of the M2 marker CD206.
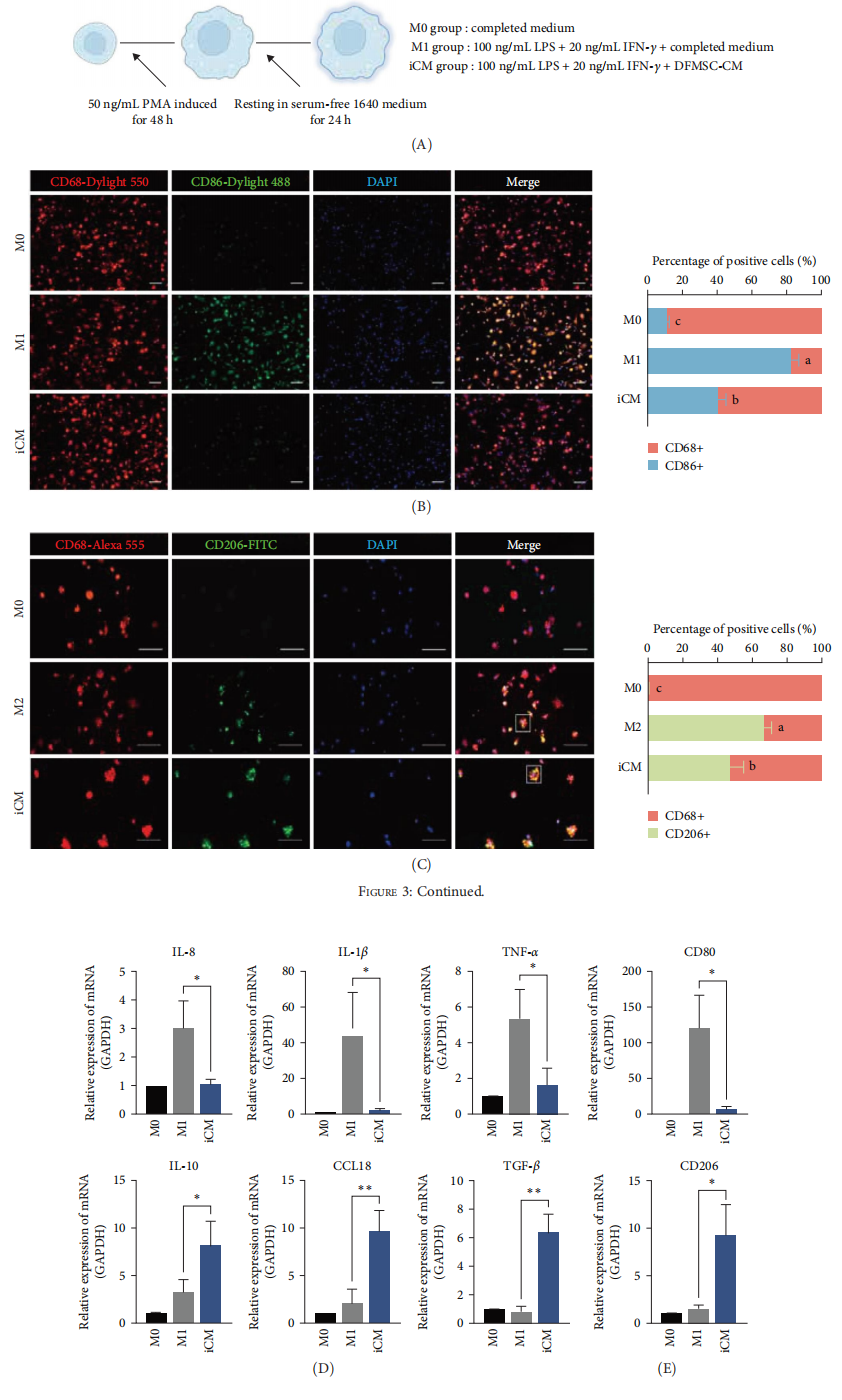
Figure 3: DFMSC-CM promotes anti-inflammatory macrophage polarization in vitro under inflammatory conditions
IV. Transcriptome Sequencing and Bioinformatics Analysis
Transcriptomic analysis revealed that differentially expressed genes between DFMSC-CM–treated and untreated macrophages were predominantly enriched in the MAPK signaling pathway. Western blot assays confirmed that DFMSC-CM markedly enhanced phosphorylation of ERK1/2 within the MAPK pathway and upregulated the expression of EGR1 protein.
Further experiments using the ERK1/2 inhibitor PD98059 to block the MAPK-ERK1/2 pathway showed a reduction in EGR1 expression, an increase in pro-inflammatory cytokine levels, and a decrease in anti-inflammatory cytokine levels. These results demonstrate that DFMSC-CM regulates macrophage polarization through the MAPK-ERK1/2-EGR1 signaling axis.
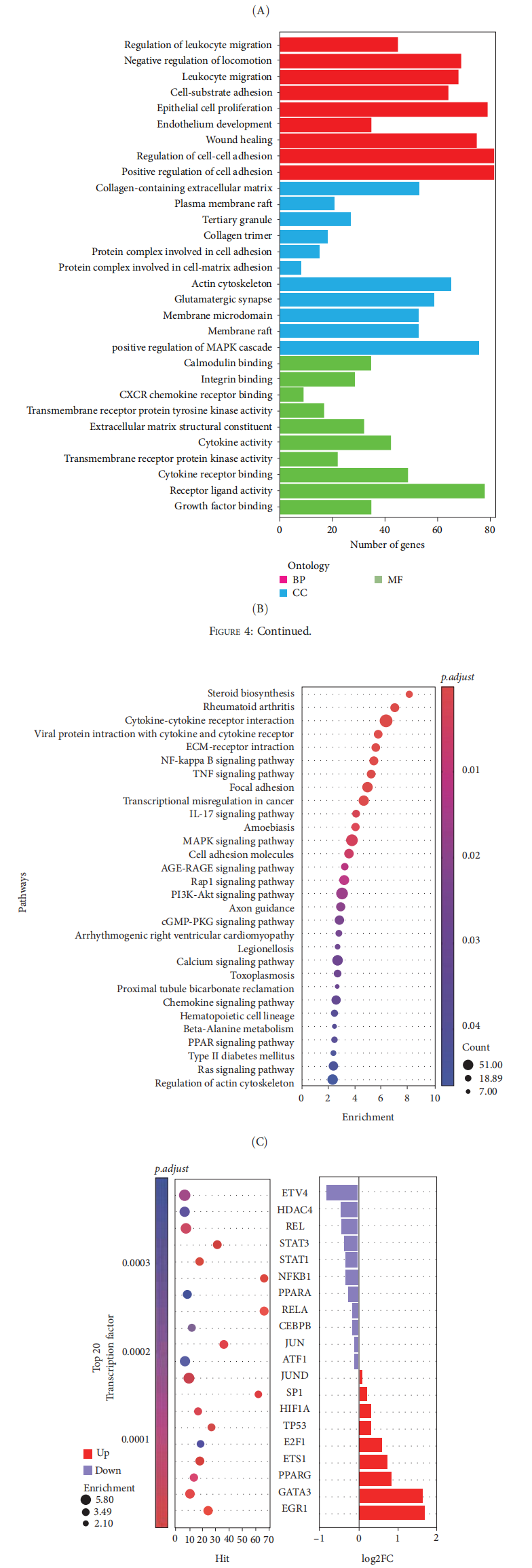
Figure 4: Transcriptomic analysis of M1 and iCM macrophages
V. Critical Role of EGR1 in Macrophage Polarization
To further validate the role of the transcription factor EGR1, the researchers used an EGR1 knockout THP-1 cell line developed by EDITGENE. The results showed that in EGR1-deficient macrophages, DFMSC-CM failed to effectively suppress pro-inflammatory cytokine expression. Additionally, the expression of the M2 marker CD206 decreased, while the M1 marker CD80 increased. These findings confirm the pivotal role of EGR1 in DFMSC-CM–mediated regulation of macrophage polarization.
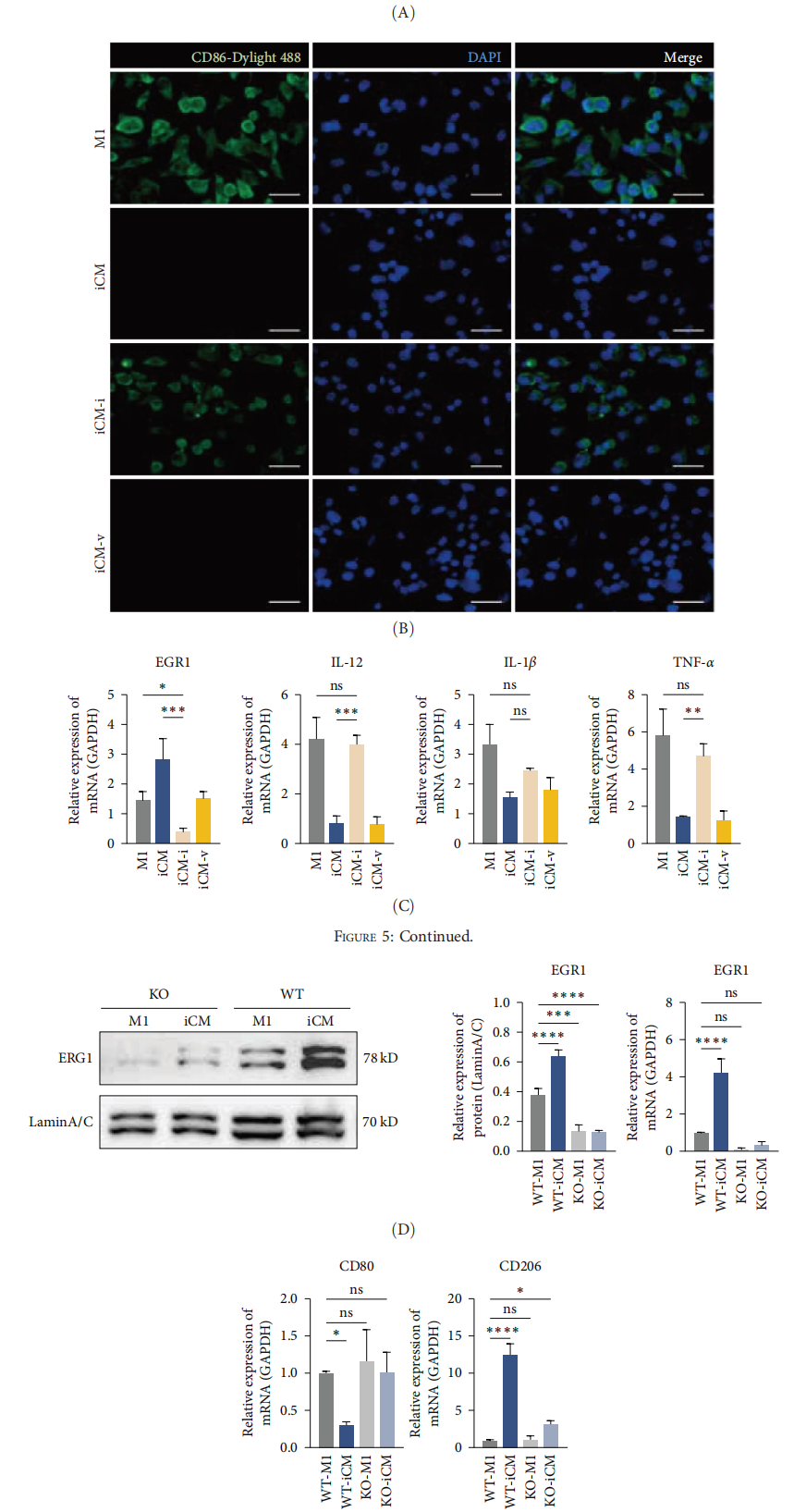
Figure 5: DFMSC-CM regulates macrophages via the MAPK-ERK1/2-EGR1 axis
Conclusion
This study uncovers the molecular mechanism by which DFMSC-CM regulates macrophage polarization through the MAPK-ERK1/2-EGR1 signaling axis, providing a scientific basis for leveraging DFMSCs and their secretome in anti-inflammatory therapies. With their broad availability, minimal ethical concerns, and potent immunomodulatory properties, DFMSCs hold great potential as a novel resource for treating inflammation-related diseases.
EDITGENE specializes in customized gene knockout cell line development, delivering powerful tools that help researchers unlock critical mechanisms and drive discoveries from bench to bedside.Ready to accelerate your gene editing research?Contact us today to tailor a solution for your study.EDITGENE– making target validation more rapid and more precise.
Recent Blogs
- 1. [Quality Share]Cracking the Delivery Puzzle: How Can Gene Editing Precisely Reach Target Cells?
- 2. [Literature Review] CRISPR–Csm: Real-time Single-Molecule Tracking of RNA Dynamics in Live Cells
- 3. [Literature Review] New Developments in Prime Editing: Phage-Assisted Evolution and Protein Engineering Yield More Efficient and Compact Prime Editors
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com
Search
Search
Recent Post
-

29 April.2025
【Literature Review】EZH2 Knockout Mouse Model Sheds Light on Ferroptosis Mechanisms in Pulpitis
-
![[Literature Review] CRISPR Screening Reveals the Critical Role of NUS1 in Prostate Cancer Survival and Growth](/uploads/20250429/st8wmjq4plYCdzgA_53c82bdd67704fe0e159246934f924ee.png)
29 April.2025
[Literature Review] CRISPR Screening Reveals the Critical Role of NUS1 in Prostate Cancer Survival and Growth
-

29 April.2025
EDITGENE Supports Research Team in Uncovering New Mechanism of Macrophage Polarization Regulated by Dental Pulp Stem Cells
















Comment (4)