[Practical Insights]Designing Donor Templates
Prime Editing

When CRISPR-Cas9 induces a double-strand break (DSB) in the genomic DNA, it simultaneously triggers the cell’s DNA repair mechanisms. To guide the repair via homology-directed repair (HDR) and introduce desired mutations or knock-in sequences, an additional donor template must be provided. This donor template typically consists of a 5’ homology arm, the mutation or knock-in sequence, and a 3’ homology arm.
The design of the donor template is critical for HDR efficiency. Below are key principles and recommendations for designing effective donor templates.
Both ssODN (single-stranded oligodeoxynucleotide) and dsDNA (plasmid DNA) can serve as donor templates, each with distinct advantages depending on the experimental context:
ssODN: Typically 120–200 bp in length and chemically synthesized, with homology arms of ~40-90 bp on each side. Despite the shorter homology regions, ssODNs often achieve high HDR efficiency in many cell types. They induce minimal cellular stress and are less likely to trigger innate immune responses, allowing higher transfection amounts to further improve HDR efficiency.
dsDNA: Better suited for long-sequence knock-ins or cases requiring extended homology arms due to complex target regions. However, dsDNA can stimulate cellular stress, as cells may recognize it as free DNA and activate apoptotic pathways, reducing the likelihood of obtaining positive clones. High intracellular dsDNA concentrations also increase the risk of random genomic integration, potentially causing insertional mutations or genome instability.
This trade-off makes ssODNs preferable for small edits or point mutations, while dsDNA is typically reserved for larger insertions.
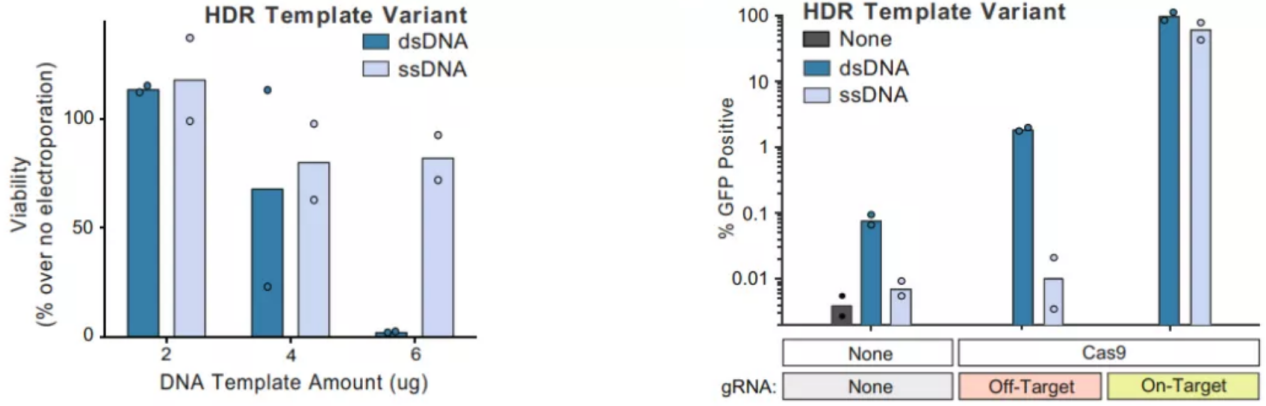
Figure 1. Comparison of dsDNA and ssDNA donor templates in terms of cytotoxicity and random integration
- Based on the characteristics of ssODN and dsDNA: For constructing point mutation cell lines, ssODN is generally preferred as the donor template. When the sequences flanking the target site are complex, the experimental design can be optimized by:
• Extending the homology arms of the ssODN to increase recombination efficiency.
• Increasing the amount of ssODN used in the transfection.
• Using plasmid DNA (dsDNA) as the donor template if necessary.
For knock-in cell lines, either ssODN or dsDNA can be used depending on the insert size: Small fragments (e.g., short tags) are suitable for ssODN. Long fragments require a plasmid-based dsDNA donor. In general, the shorter the knock-in fragment, the higher the HDR efficiency, as the length of the inserted sequence is inversely correlated with homologous recombination efficiency.
The length and homology of the arms are critical for efficient HDR. Typically, each homology arm ranges from 500-1000 bp, and for larger insert fragments, longer homology arms can be used to improve knock-in efficiency.
When choosing templates for the homology arms, it is recommended to amplify from the genome of the target cell line. This avoids sequence differences between donor and host genomes that could reduce HDR efficiency.
After HDR repair, if the target site still retains the full gRNA recognition sequence and PAM, Cas9 may cut again, potentially generating undesired mutations. To prevent this, silent mutations can be introduced in the PAM or adjacent sequences within the donor template to make the site resistant to Cas9 recognition.
- For knock-in donor templates, codon optimization can be applied. By adjusting the codon usage in the exogenous gene to better match the translation machinery of the host cell, this strategy can improve expression efficiency and accuracy of the inserted gene.
- In general, when using ssODN as the donor template, selection markers are not introduced. This is because ssODNs have limited capacity and their HDR efficiency is usually high enough that subsequent screening is straightforward.
When using plasmid DNA as the donor template, selection markers can be included to facilitate the identification and expansion of positive clones. In addition to antibiotic resistance genes, fluorescent reporter genes can also be incorporated to enable visual screening. However, it is important to consider that the length of exogenous sequences may impact HDR efficiency.













![[Practical Insights]Designing Donor Templates](/uploads/20250328/ESzk5OC49wpxIHVv_3cbfa5e98ea1d238127fe23c72b0f4b2.png)

Comment (4)