Protein structure is fundamental to biological function. Encoded by diverse DNA sequences, polypeptides undergo splicing, modification, and folding to form proteins with highly specific three-dimensional architectures.
Importantly, even a single protein can adopt multiple conformations under different conditions, enabling it to perform distinct biological tasks. Understanding these structural states is therefore key to uncovering protein function.
Over the past decade, advances in cryo-electron microscopy (cryo-EM) have enabled researchers to capture protein structures under near-physiological conditions, offering unprecedented insight into protein dynamics and conformational transitions.
As one of the most widely used tools in genome editing, the Cas protein family and Cas9 in particular, has become a major focus of structural and functional research. In this article, we take a closer look at the bilobed architecture of Cas9 and examine how its inactive Apo-Cas9 conformation contributes to functional regulation.
Bilobed Architecture of the Cas9 Protein
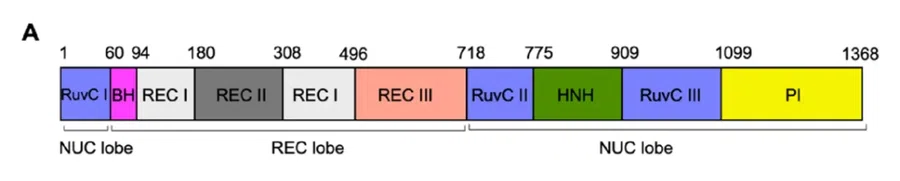
The Cas9 protein adopts a characteristic bilobed architecture, consisting of two major structural regions: the Recognition (REC) lobe and the Nuclease (NUC) lobe.
The REC lobe is composed of three α-helical domains—REC I, REC II, and REC III—and plays a central role in sensing and verifying correct base pairing between the sgRNA (or crRNA) and the target DNA. Notably, the REC lobe exhibits no significant homology to other known proteins, highlighting its unique role in nucleic acid recognition.
In contrast, the NUC lobe contains the catalytic machinery responsible for DNA cleavage. It includes the conserved HNH domain and a RuvC-like domain, which is split into three segments: RuvC I, RuvC II, and RuvC III. Using Streptococcus pyogenes Cas9 (spCas9) as an example (Figure A), the RuvC-like domain is structurally interrupted by the REC lobe and the HNH domain.
RuvC I is located at the N-terminus and connects to the REC lobe via an arginine-rich Bridge Helix (BH).
RuvC II lies at the C-terminus of the REC lobe, followed sequentially by the HNH domain and RuvC III.
Extending from RuvC III, the PAM-interacting (PI) domain occupies the extreme C-terminus of Cas9 and is responsible for PAM recognition.
In three-dimensional space, folding of the polypeptide chain brings the separated RuvC segments together to form an integrated catalytic pocket. This split RuvC-like architecture is a distinctive feature of Cas9, differing from classical prokaryotic RuvC enzymes that form a continuous, compact globular domain.
By separating the RuvC segments with the REC and HNH domains, Cas9 enables tight regulatory control over nuclease activity. This structural arrangement allows Cas9 to switch flexibly between inactive and active conformations, ensuring precise and programmable DNA cleavage.
Structural Features of Apo-Cas9
Genome editing activity requires formation of the sgRNA–Cas9 complex. In the absence of sgRNA, Cas9 remains catalytically inactive, a conformation known as the apo state. The term “apo” originates from the Greek word ἀπό, meaning “away from,” and in biochemistry refers to a protein that lacks its functional ligands, such as substrates or cofactors.
Cryo-EM studies reveal that in the Apo-Cas9 state, the REC and NUC lobes adopt a relatively closed conformation, and the nucleic acid-binding groove between them remains inaccessible.
The REC subdomains are locked in an inactive arrangement, while the bridge helix maintains a relatively straight configuration. Positively charged surfaces that would normally interact with negatively charged DNA are not exposed.
In addition, the three RuvC segments fail to assemble into a functional catalytic pocket, and the HNH domain is positioned away from the cleavage site.
The PI domain appears structurally unstable, exhibiting diffuse electron density consistent with a flexible, mobile “tail.” Collectively, these structural features explain why Apo-Cas9 lacks DNA cleavage activity.
Regulatory Role of the REC Lobe in the Apo State
Although Apo-Cas9 is catalytically inactive, recent studies suggest it is not entirely functionless. Research from the University of Michigan demonstrated that Neisseria meningitidis Cas9 (NmeCas9) can regulate viral spacer acquisition efficiency in bacterial adaptive immunity in response to crRNA abundance.
Through systematic deletion of individual structural domains, the study revealed that while the NUC lobe participates directly in spacer acquisition, the REC lobe plays a key regulatory role by sensing crRNA levels.
These findings highlight the REC lobe as a critical regulatory element, extending its function beyond target recognition to broader control of Cas9 activity.
As both a cornerstone of bacterial adaptive immunity and a powerful genome-editing tool, Cas9 remains a central focus of life science and biotechnology research. Its bilobed architecture and conformational flexibility enable precise regulation of DNA cleavage, underpinning its versatility and reliability in CRISPR-based applications.
With over a decade of expertise in CRISPR-Cas systems, EDITGENE provides high-quality Cas enzymes supported by a robust protein purification platform.
EDITGENE also offers stable Cas9-expressing cell lines with high expression levels and editing efficiency, supporting applications such as knockout cell line generation and sgRNA library screening. These tools are designed to streamline workflows, accelerate research timelines, and support successful genome-editing outcomes.
References
1. Zhou, Xufei et al. “Cas9 senses CRISPR RNA abundance to regulate CRISPR spacer acquisition.” Nature vol. 647,8091 (2025): 1054-1062. doi:10.1038/s41586-025-09577-9
2. Babu, Kesavan et al. “Coordinated Actions of Cas9 HNH and RuvC Nuclease Domains Are Regulated by the Bridge Helix and the Target DNA Sequence.” Biochemistry vol. 60,49 (2021): 3783-3800. doi:10.1021/acs.biochem.1c00354
Follow us on social media





Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com



















