[Literature Review] CRISPR-GPT — Your AI Gene Editing Assistant
Gene Editing
 Large Language Models (LLMs) have demonstrated remarkable language-processing capabilities in fields such as mathematics and chemistry. However, in biomedicine, due to the lack of domain-specific knowledge and the inherent complexity of biological systems, general-purpose LLMs (e.g., ChatGPT) often suffer from “hallucination” issues, making it difficult to provide researchers with accurate gene-editing experimental design solutions.
Large Language Models (LLMs) have demonstrated remarkable language-processing capabilities in fields such as mathematics and chemistry. However, in biomedicine, due to the lack of domain-specific knowledge and the inherent complexity of biological systems, general-purpose LLMs (e.g., ChatGPT) often suffer from “hallucination” issues, making it difficult to provide researchers with accurate gene-editing experimental design solutions.
To address this challenge, the teams led by Prof. Le Cong at Stanford University and Prof. Mengdi Wang at Princeton University, in collaboration with Google DeepMind, published a study in Nature Biomedical Engineering titled “CRISPR-GPT for agentic automation of gene-editing experiments.” This work reports the development of the world’s first LLM-based system for automated gene-editing experiment design—CRISPR-GPT.

Original link: https://www.nature.com/articles/s41551-025-01463-z
Spotlight
1. LLMs Empower Experimental Design
A multi-agent system was developed to automate the entire workflow from experiment planning to data analysis, with seamless integration with databases such as CRISPick.
2. Three Intelligent Interaction Modes
The CRISPR-GPT system offers Meta, Auto, and Q&A modes, supporting beginner-guided operations, expert-customized workflows, and real-time query responses, respectively.
3. Simulating Expert Reasoning
By integrating CRISPR discussion data and key literature, and fine-tuning a dedicated model, CRISPR-Llama3, the system can provide accurate, logically coherent answers that mimic expert reasoning.
1. Meta Mode
Designed for beginners with limited gene-editing experience, this mode provides step-by-step guidance through the entire workflow, from CRISPR system selection to data analysis. Key tasks—including delivery method planning, gRNA design, and off-target assessment—are fully covered, enabling newcomers to complete a comprehensive gene-editing experiment design.
2. Auto Mode
Suitable for researchers with some gene-editing experience, this mode automatically decomposes tasks and generates personalized workflows based on user-submitted requirements, addressing specific experimental needs without starting from scratch.
3. Q&A Mode
In this mode, users can consult the system at any time with gene-editing-related questions, receiving real-time, expert-level responses.
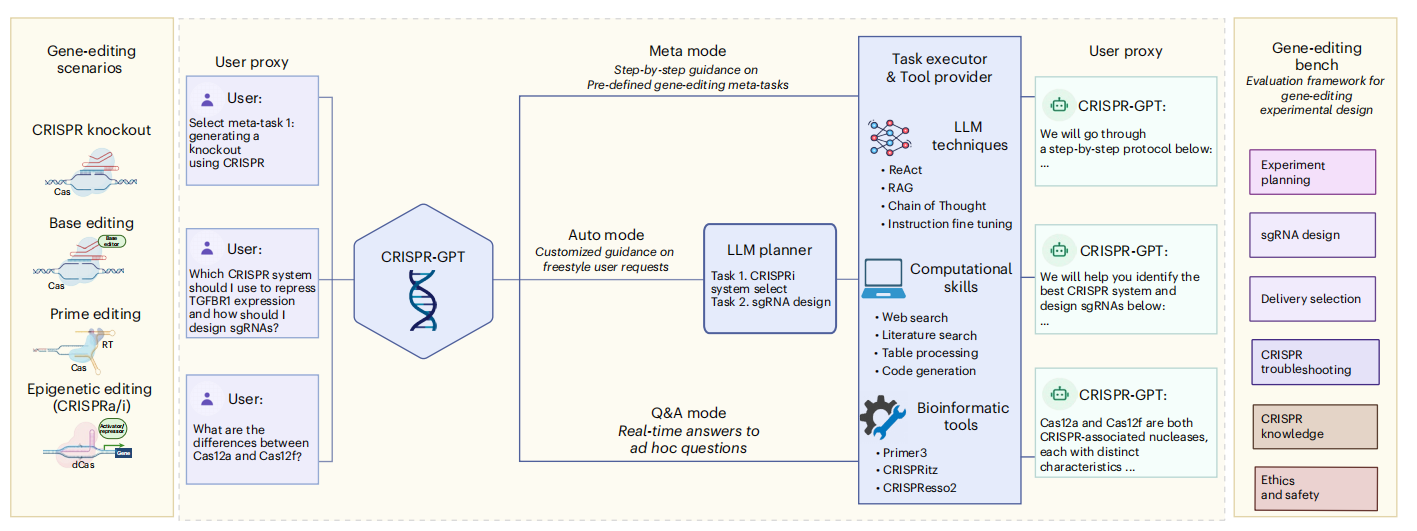
Figure 1. Overview of CRISPR-GPT
1. Experimental Planning
The task-planning agent within CRISPR-GPT can decompose user-submitted requirements into a chain of tasks through multimodal interactions, enabling the automatic construction of personalized experimental workflows.
Evaluation results show that CRISPR-GPT significantly outperforms general-purpose LLMs in experimental planning accuracy. Its GPT-4o-based version (GPT-4 optimized) achieves accuracy and F1 scores above 0.99 (Figure 2b), demonstrating strong capabilities in workflow comprehension and task management.
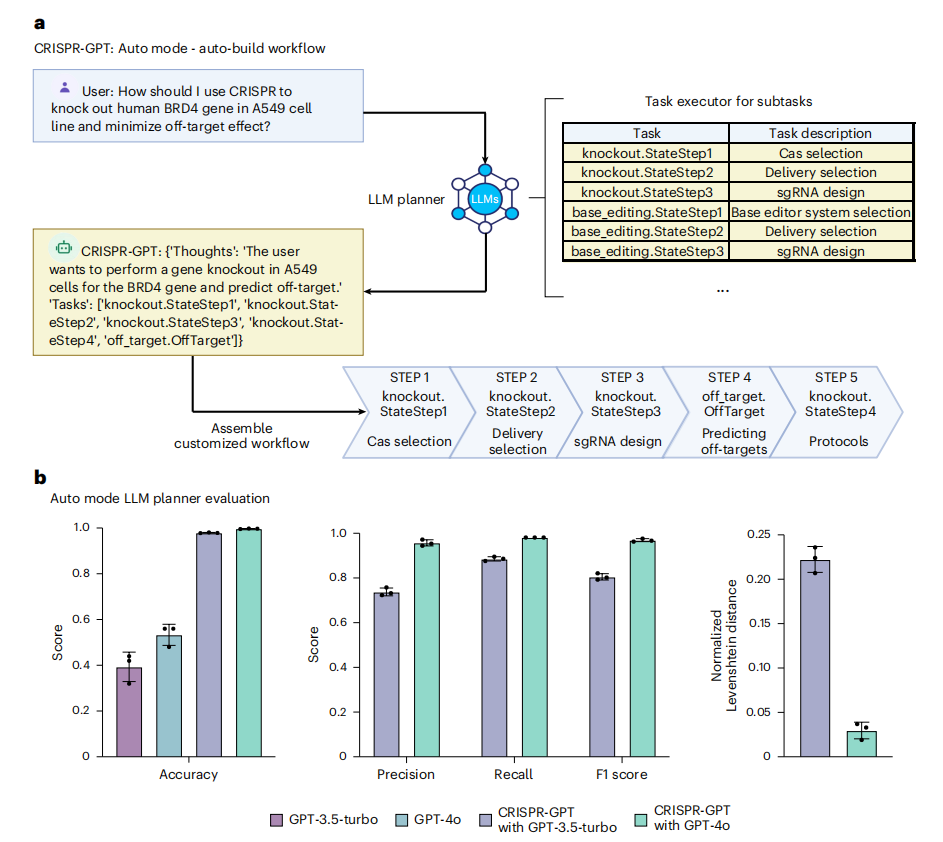
Figure 2. Task decomposition and experimental planning in CRISPR-GPT auto mode, with performance evaluation
2. Delivery Method Selection
The agent identifies specific biological keywords in user requests and combines Google search with literature retrieval to recommend optimal delivery strategies. In tests across 50 biological systems, CRISPR-GPT outperformed GPT-4 and GPT-3 models, showing a clear advantage in complex tasks such as hard-to-transfect cell lines and primary cells (Figure 3b).
3. gRNA Design
By integrating authoritative databases such as CRISPick and leveraging the semantic reasoning capabilities of large language models (LLMs), CRISPR-GPT employs a chain-of-tables approach to efficiently select gRNAs under specific experimental requirements (Figure 3c).
Test results for gRNA design tasks indicate that CRISPR-GPT significantly outperforms baseline models in key metrics, including target selection accuracy and parameter configuration rationality (p < 0.01) (Figure 3d).
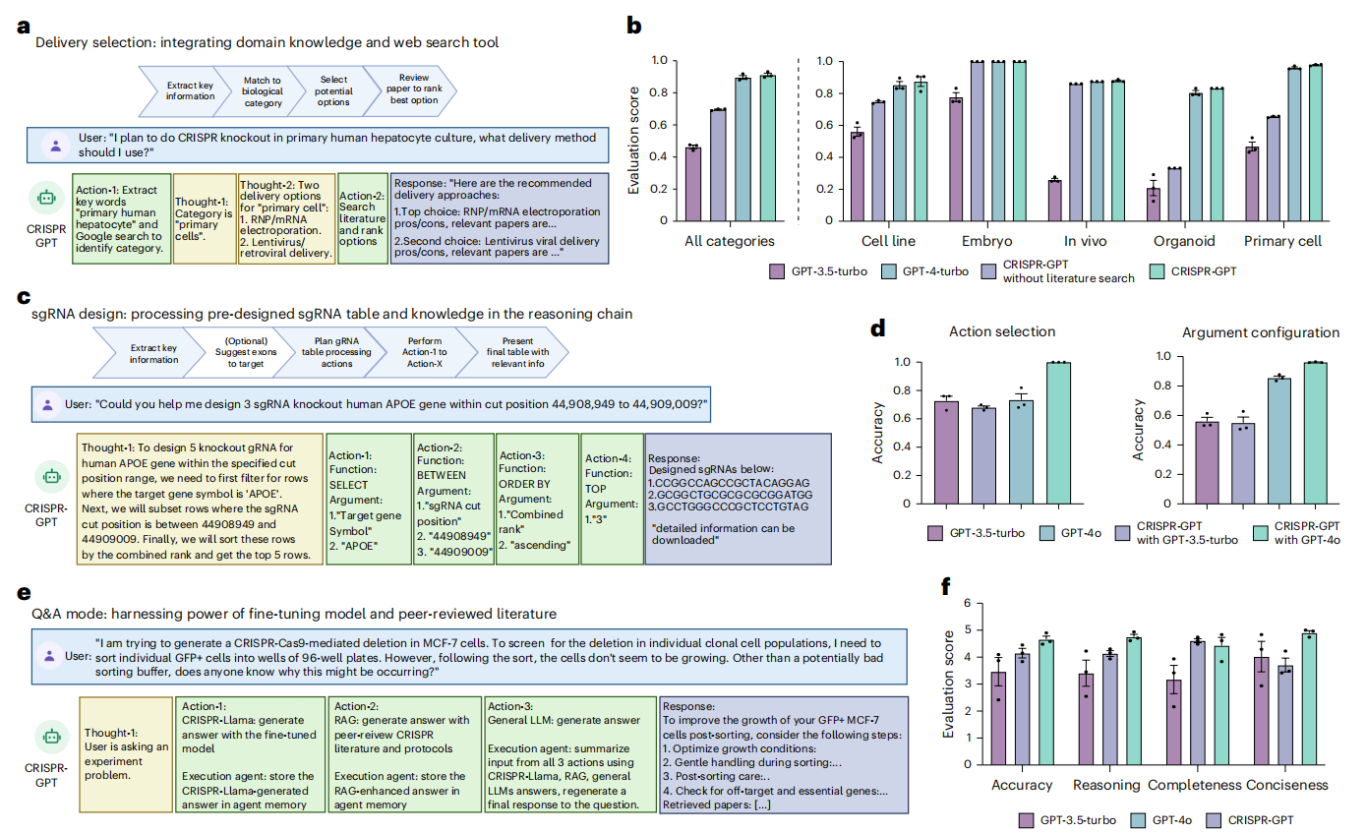
Figure 3. CRISPR-GPT enables automation of gene-editing research and experimental tasks
1. Multi-Gene Knockout Experiment
The researchers used CRISPR-GPT to design knockout strategies targeting four tumor-related genes in the lung adenocarcinoma cell line A549. The experimental results showed an editing efficiency of 80% (Figure 4b).
2. Epigenetic Editing
In an epigenetic editing experiment targeting the melanoma A375 cell line, the researchers followed CRISPR-GPT guidance to select a CRISPR activation system, design three dCas9 gRNAs, and perform validation workflows. Post-editing protein expression analysis revealed activation efficiencies of 56.5% and 90.2% for NCR3LG1 and CEACAM1, two genes associated with cancer immune tolerance (Figure 4f).
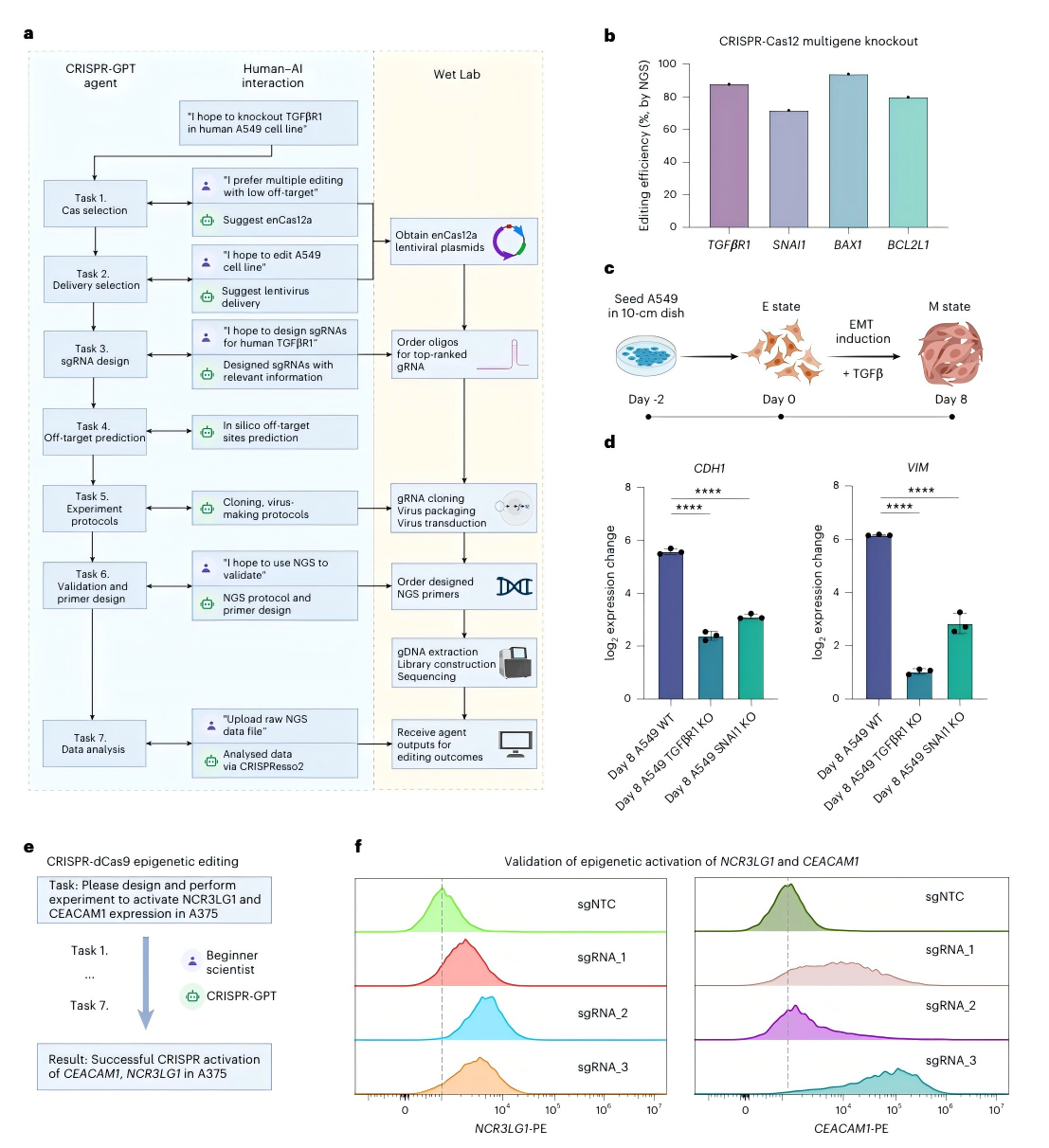
Figure 4. Experimental validation of CRISPR-GPT in knockout and activation experiments
In Conclusion, CRISPR-GPT enables automated design of gene-editing experiments, providing researchers with intelligent solutions that maintain editing precision while reducing operational complexity and enhancing reproducibility.
Looking forward, by integrating with automated laboratory platforms and robotic systems, CRISPR-GPT has the potential to achieve end-to-end automation—from computational design and data analysis to physical execution—accelerating applications of gene editing in areas such as agricultural breeding and disease therapy.















![[Literature Review] CRISPR-GPT — Your AI Gene Editing Assistant](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)